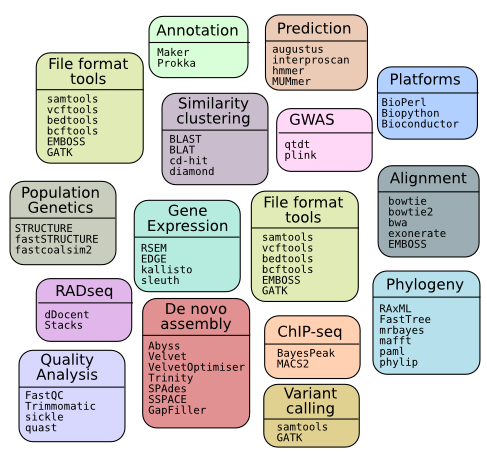
Available Software
Contents
Introduction
We now highly recommend conda to install your packages:
Conda. THIS IS THE FUTURE. We highly recommend this
We are testing Bioconda on Marvin on a per user basis, so you have total control over it.
1) Log in and from the command line type (only do this once!):
install-bioconda
2) Either log out and back in or type:
source ~/.bashrc
3) We strongly advise using environments: https://conda.io/docs/user-guide/tasks/manage-environments.html . You can do this in many ways. Please see the link for more details (here you can specifiy eact version etc ..). The easiest usage would be: e.g. conda create -n NAME_OF_ENV PACKAGE_TO_INSTALL (pick a package you are interested in from here: installing packages: https://bioconda.github.io/recipes.html ). Roary is a package name as an example.
conda create -n roary roary
conda activate roary
conda update roary
You are now ready to use this package.
Conda deactivate to leave this environment.
This works for me and a test user. But may fail for you, so be patient with us.
To list all the environments you have created:
conda info –envs
installing packages:
https://bioconda.github.io/recipes.html
module load approach
To see all the available software, the command to type is:
moduleav
This is a fast wrapper command for the underlying slower:
module available
Categorised Listing
Raw Listing 07 Dec 2016
--------------------------------- /usr/local/Modules/versions ----------------------------------------------------- 3.2.10 ---------------------------- /usr/local/Modules/3.2.10/modulefiles ------------------------------------------------ AdapterRemoval/gitv0_abe4a3e(default) ArtificialFastqGenerator/1.0.0(default) AsperaConnect/3.6.2 BAPS/6.0 BUSCO/1.22 BamQC/gitv0_480c091 BayeScEnv/1.0 BayeScEnv/gitv0_beb60d2(default) BayeScan/2.1(default) CAFE/3.1 CEGMA/2.5(default) CRISP/2 CRISP/5 CRISP/gitv0_7bfcd34(default) EIGENSOFT/6.0.1(default) EMBOSS/6.6.0(default) FASTQC/0.11.5(default) FastML/3.1 FastML/gitv0_bb5d761 FastTree/2.1.8(default) GapFiller/v1-10 GeneMark_ES/3.48(default) HTSlib/1.3 HTSlib/1.3.1(default) HTSlib/gitv1_0de7fe5 MCR/v713 MUMmer/3.23 MUMmer/3.23b(default) Mash/gitv0_2edc42 Mash/v1.1.1(default) Mauve/2015-02-13 NGSToolsApp/2.1.5(default) OpenBLAS/0.2.14 OpenBLAS/0.2.18 OpenBLAS/0.2.19(default) OrthoFinder/0.6.0 PAGIT/1(default) PEAR/gitv0_a37a84b(default) PEAT/gitv0_27981d7(default) PGAP/1.2.1 PGDSpider/2.0.8.2(default) Prodigal/gitv0_004218f R/3.2.1(default) R/3.2.3 R/3.3.1 RAxML/gitv0_4692545(default) RSEM/1.2.25(default) RSEM/1.2.27 SPAdes/3.9.0(default) SSPACE/3.0 STAR/2.4.2a(default) T-COFFEE/11.00.8 VarScan/2.3.7(default) VelvetOptimiser/2.2.5(default) abacas/1.3.1(default) abyss/1.9.0 act/v1 allpathslg/52488 angsd/gitv0_212d58 angsd/gitv1_89d8fa9(default) arlequin/3.5.2(default) artemis/16.0.0(default) assembly-stats/gitv0_003a372 augustus/3.0.3 augustus/3.2.2(default) bamtools/gitv0_02c2be8(default) bcftools/gitv0_47e811c bedtools/2.23.0 bedtools/2.25.0(default) bioawk/gitv0_5e8b41d(default) blastScripts/1.0.0(default) blastScripts/1.0.1 blastall/2.2.26(default) blat/35 boost/1.60.0(default) boost/1.61.0 bowtie/1.0.0 bowtie2/2.1.0(default) bowtie2/2.2.4 bwa/0.7.12(default) bwa/0.7.7 bzip2/1.0.6 capnproto/0.5.3 cd-hit/gitv0_5acf038(default) clustalx/2.1 crimap/2.504(default) cufflinks/2.2.1(default) dDocent/gitv0_7dea314 diamond/gitv0_39f2a71 dot ea-utils/gitv0_88c9ad5 ensembl-git-tools/gitv0_68ac669(default) ensembl/gitv0_bb48912(default) exonerate/2.2.0(default) fastSTRUCTURE/1.0 fastSTRUCTURE/190315(default) fastsimcoal2/2.5.2.21 fastsimcoal2/2.5.2.8(default) freebayes/gitv0_dbb6160 gatk/3.4.0 gatk/3.5.0 gatk/3.6.0(default) gcc/4.9.3 geneid/1.4.4(default) general_script_tools/(default) gnuparallel/20160422 gsea/2.2.2(default) gsl/2.1 gubbins/gitv0_fe094b0 gubbins/gitv1_8a12ae7(default) gwas harvest-tools/gitv0_ebf7ecc hdf5/1.8.15-patch1(default) hdf5/1.8.16 hmmer/2.3.2 hmmer/3.1b1 hmmer/3.1b2(default) interproscan/5.4-47.0(default) kallisto/git0_f0678a2 lamarc/2.1.10(default) lpsolve/5.5.2.3 mafft/7.147(default) maker/2.31.8(default) masurca/2.3.2(default) mcl/14-137(default) mdb/0.6(default) merlin/1.1.2(default) module-info modules mpifast/gitrev2(default) mrbayes/3.2.6 muscle/3.7 newick_utils/4.9.3 newick_utils/gitv0_614bf6c(default) ngs/gitv0_aef5f50 null openmpi/1.10.2 openmpi/1.6.5 openmpi/2.0.0(default) oraclejava/jdk1.8.0_74 paml/4.7a(default) parsnp/gitv0_63bea13 pblat/1.6.0(default) perl/5.20.2(default) perl/5.24.0 phylip/3.696(default) phyml/gitv0_0f6d745 picard-tools/1.119 picard-tools/2.1.0 picard-tools/2.1.1 picard-tools/2.7.1(default) prank-msa/gitv0_3e44771(default) prokka/1.11 prokka/gitv1_8f07048(default) prosite/1.79.1 protobuf/gitv0_0622030 psmc/gitv0_e5f7df5(default) pyrad/gitv0_704ffbf python/2.7(default) python/2.7.12 python/3.4 qtdt/2.6.1(default) quast/4.1 rainbow/2.0.4 rapsearch/gitv0_95c866e(default) readseq/2.1.30(default) repeatmasker/4.0.5(default) rmblastn/2.2.28(default) rnammer/1.2(default) roary/gitv0_c4d9b77(default) samtools/0.1.18 samtools/0.1.19(default) samtools/0.1.19b samtools/1.2 samtools/1.3.1 scalaBLAST/2.0(default) scipio/1.4.1(default) script_tools/1.0(default) seqtk/gitv0_1a8319b(default) sickle/gitv0_f3d6ae3(default) signalp/4.1(default) smalt/0.7.6(default) snap_alignment/1.0beta.17(default) snap_hmm/11_29_2013(default) snape-pooled/31(default) snippy/gitv0_848a62e snpcallphylo/0.7 sparsehash/gitv0_4cb9240 sra-tools/gitv0_814d6f7(default) srst2/gitv1_e30dd01(default) srst2/gitv2_41b3f46 stacks/1.35 stacks/1.37 stacks/1.40(default) stacks/1.41 stampy/1.0.23(default) structure/2.3.4(default) tRNAscan-SE/1.3.1(default) tabix/gitv0_1ae158a tabtk/gitv0_741f3f0 tophat/2.0.10(default) treemix/gitv0_23c15a2(default) trimmomatic/0.32 trinity/2.2.0(default) trinity/20140717 use.own vcflib/gitv0_e3ab177 vcftools/0.1.11 vcftools/0.1.12a vcftools/gitv0_4fce995(default) vdb/gitv0_f1497a1 velvet/1.2.10(default) velvet/gitv0_9adf09f vsearch/gitv0_174f810(default)