Mapping to Reference Talk
Mapping to a reference genome
Contents
- 1 Contents
- 2 The mapping process
- 3 Gettng a Reference sequence
- 4 Mapping is a vital step
- 5 NGS data - Challenges
- 6 Mapping process considerations
- 7 Mapping process considerations
- 8 Mapping algorithms
- 9 Mapping output
- 10 Mapping output
- 11 SAM/BAM format: header section
- 12 SAM/BAM format: alignment section
- 13 SAM/BAM format: FLAG
- 14 SAM/BAM format: CIGAR
- 15 SAM/BAM format: CIGAR example
- 16 SAM/BAM format: optional tags
- 17 SAM/BAM format: option tags
- 18 SAM/BAM format
- 19 SAM/BAM format: SAM parser
- 20 Tophat2
Contents
- Overview
- Mapping process: Algorithms and tools
- Mapping output: SAM/BAM specification
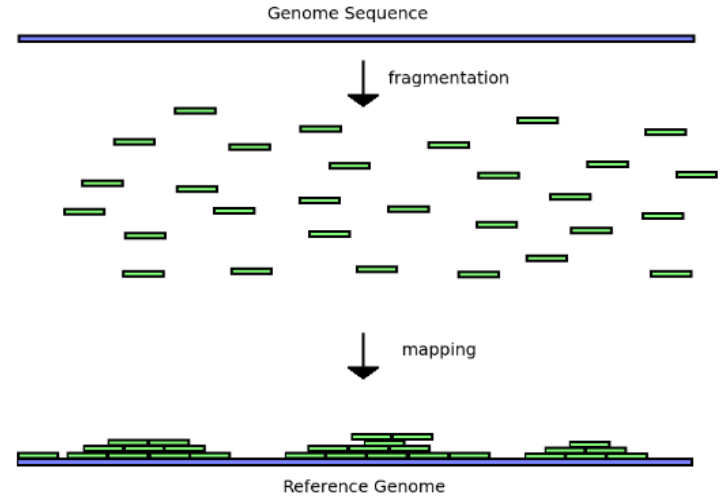
The mapping process
Gettng a Reference sequence
- A reference is a consensus sequence, built up from high quality sequencing samples. This can be genome or a transcriptome.
- This should be in fasta format.
Mapping is a vital step
NGS data - Challenges
- Massive Data:
- Illumina Hiseq 2500: 160GB in 2x150bp reads
- Natural variability: SNPs, indels, de novo
mutations, CNVs
- Sequencing errors
- RNA-seq: splice junctions to be considered
- Computing resources
Mapping to a reference genome
U Trivedi 2016-05-19
6
Mapping process considerations
Different mappers depending on:
Read length SNVs? Indels? DNA or RNA Single end or paired end? Should multiple hits be allowed?
Which mapper to use? Mapping to a reference genome
U Trivedi 2016-05-19
7
Mapping process considerations
Mapping to a reference genome
U Trivedi 2016-05-19
8
Mapping algorithms
- BLAST
- Allows comparing and searching amino-acid and DNA
sequences in a database of sequences
- Uses a heuristic algorithm: cannot guarantee the
optimal alignment
- Too slow for NGS
- Based on Hashes
- High memory footprint
- Slow for NGS
- Burrows Wheeler Transform
- Very fast and low memory footprint
- Very sensitive to errors
- Hybrids
Mapping to a reference genome
U Trivedi 2016-05-19
9
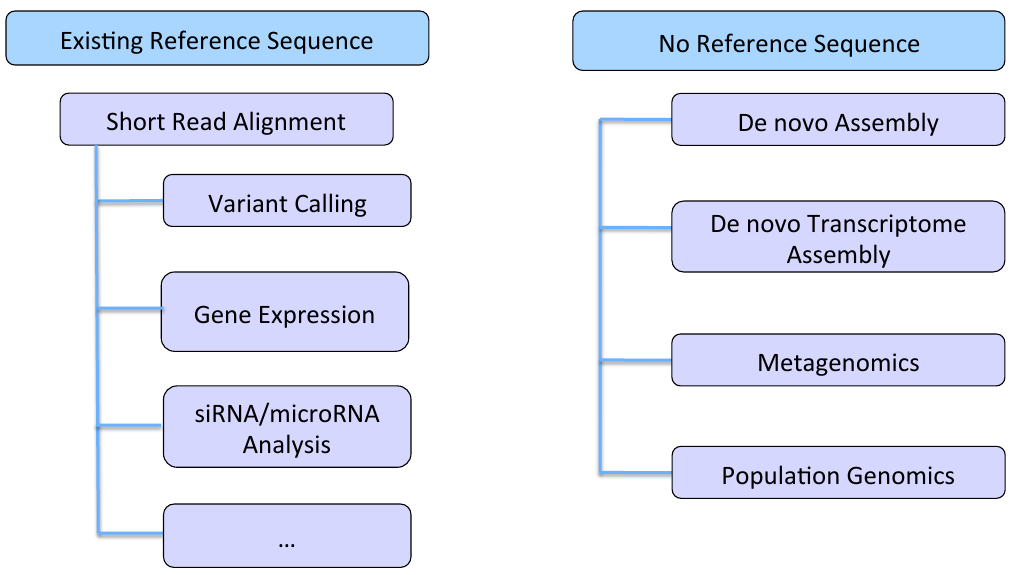
Mapping output
SAM/BAM format http://samtools.github.io/hts-specs/SAMv1.pdf Suppose we have the following alignment
Mapping to a reference genome
U Trivedi 2016-05-19
10
Mapping output
Corresponding SAM format will be:
Mapping to a reference genome
U Trivedi 2016-05-19
11
SAM/BAM format: header section
Mapping to a reference genome
U Trivedi 2016-05-19
12
SAM/BAM format: alignment section
Mapping to a reference genome
U Trivedi 2016-05-19
13
SAM/BAM format: FLAG
e.g. 1059, what does this mean? http://broadinstitute.github.io/picard/explain-flags.html Mapping to a reference genome
U Trivedi 2016-05-19
14
SAM/BAM format: CIGAR
- The CIGAR string is a sequence of base lengths with an associated
operation.
- Used to indicate things like which bases align (either a match/
mismatch) with the reference, are deleted from the reference, and are insertions that are not in the reference.
Mapping to a reference genome
U Trivedi 2016-05-19
15
SAM/BAM format: CIGAR example
Alignment
CIGAR
Mapping to a reference genome
U Trivedi 2016-05-19
16
SAM/BAM format: optional tags
Mapping to a reference genome
U Trivedi 2016-05-19
17
SAM/BAM format: option tags
[Image]
SAM/BAM format
BAM format and BAM index
- BAM format
- - BAM format is the binary (compressed)
representation of a SAM file
- - A BAM file is smaller than its corresponding SAM file,
and can be read faster, but the content is the same
- BAM index
- - Indexing a bam allows to access the alignments from a specified region.
- - Required for alignment visualization soeware like IGV.
SAM/BAM format: SAM parser
- Samtools (http://samtools.sourceforge.net/)
- - Written in C (fast)
- - Provide various utilities for manipulating alignments in the SAM format
- SAM to BAM conversion
- Sorting (by coordinates or query-name)
- Merging several files
- BAM index
- Picard (Java)
- Pysam (Python)
Tophat2
- First published 2009, up to that point, aligners behaved like so:
- - "whenever an RNA-Seq read spans an exon boundary, part of the read will not map contiguously to the reference, which causes the mapping procedure to fail for that read."
- Splicing-aware, finds annotated and novel junctions
- Reaching end-0f-life ... handing over to HISAT2