Difference between revisions of "Functional Analysis Talk"
(Created page with "= Why do functional analysis? = * Statistical significance is not the same as biological significance * Variety of methods available to compare functional annotation to gene...") |
(No difference)
|
Revision as of 22:28, 11 May 2017
Contents
- 1 Why do functional analysis?
- 2 Functional analysis methods
- 3 Functional analysis methods
- 4 Gene Set Enrichment Analysis
- 5 Gene Set Enrichment Analysis
- 6 Gene Set Enrichment Analysis
- 7 Gene Set Enrichment Analysis
- 8 Gene Set Enrichment Analysis
- 9 Gene Set Enrichment Analysis
- 10 Gene Set Enrichment Analysis
- 11 Gene Set Enrichment Analysis
- 12 Conclusions
- 13 Further reading
Why do functional analysis?
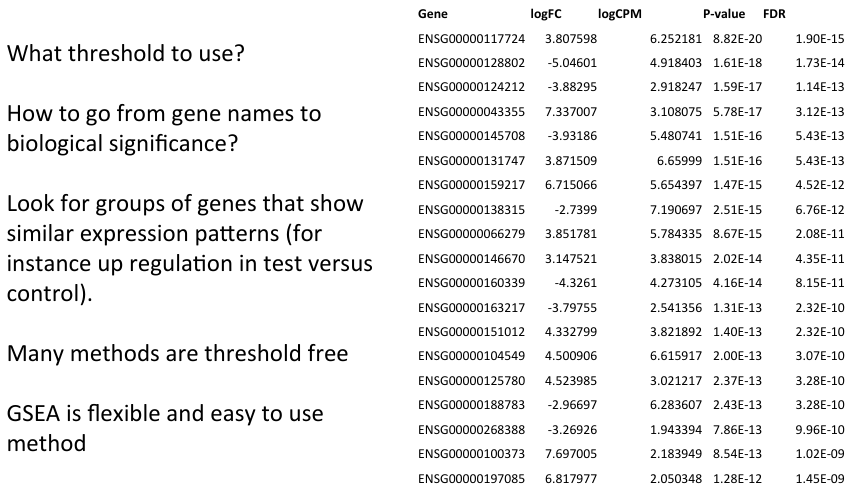
- Statistical significance is not the same as biological significance
- Variety of methods available to compare functional annotation to gene data
- General aim is to identify gene functions/categories that show an interesting expression profile
- Potentially identify genes with smaller but biologically significant changes.
Functional analysis methods
- Positional Gene Enrichment – looks for regions of chromosome showing changed expression.
- The Ingenuity Pathways Analysis (IPA)
- Several methods use Gene Ontology (GO) terms or other "gene sets"
- GO is the means by which we identify function.
- Gene Set Enrichment Analysis (GSEA)
- GOAL: Gene Ontology AnaLyzer
- GOrilla
Functional analysis methods
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis
[[File::gseawind.png]] Example GSEA report of a gene set found to be enriched among down regulated genes in cancer samples
Conclusions
- A wide variety of methods are available for functional analysis of expression data
- Aids biological interpretation of the data
- Different types of annotation can be compared to expression data.
- Many methods do not require user specified thresholds
Further reading
- Eden et al "GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene" BMC Bioinformatics 2009
- Volinia et al "GOAL: a software tool for assessing biological significance of genes groups" Nucleic Acids Res 2004
- Tamayo, et al."Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles" PNAS 2005
- Preter et al "Positional gene enrichment analysis of gene sets for high-resolution of overrepresented chromosomal regions" Nucleic Acid Research 2008
- http://www.ingenuity.com/products/ipa