Difference between revisions of "Quality of Mapping Talk"
(Created page with "= Mapping quality control = Some issues are only detectable in the context of the genome: * Duplicate reads * Fragment size distribution * Gene coverage * Completeness of data...") |
|||
| Line 50: | Line 50: | ||
= Completeness of data = | = Completeness of data = | ||
| − | {| | + | {|style="width:90%" |
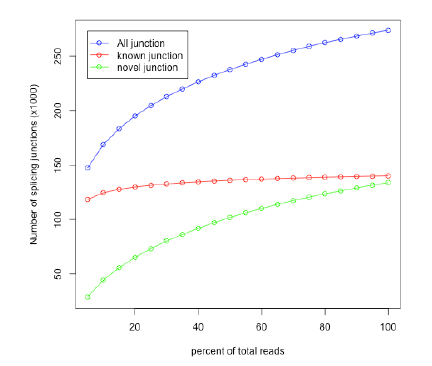
| * From a saturated RNASeq dataset, | | * From a saturated RNASeq dataset, | ||
all known splice junctions should be | all known splice junctions should be | ||
Revision as of 12:20, 9 May 2017
Contents
Mapping quality control
Some issues are only detectable in the context of the genome:
- Duplicate reads
- Fragment size distribution
- Gene coverage
- Completeness of data
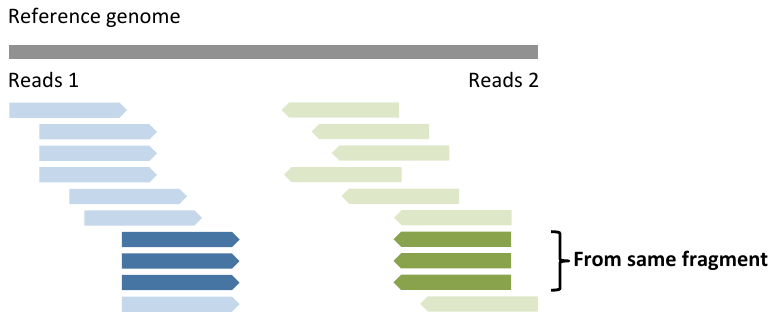
Duplicate reads
- Only detectable with paired end reads
Duplicate reads 2
- Duplicates can be PCR artefacts
- Duplicates can be real, from highly expressed transcripts
- For RNA-seq, removing duplicates is still being debated
- We don’t remove them, but it’s important to:
- - assess the duplicate rate
- - determine whether the duplicate rate can be explained by a few highly expressed genes
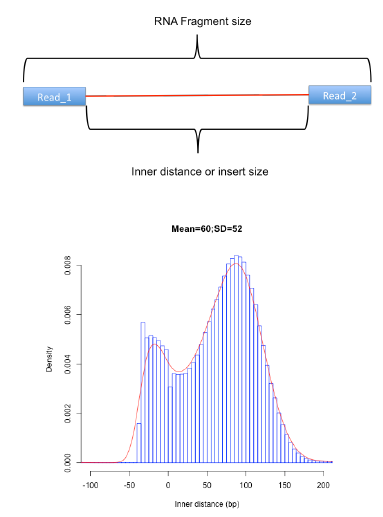
Fragment size distribution
| * Should correspond with
fragment size selected during library preparation
reads can span introns when calculating fragment size |
|
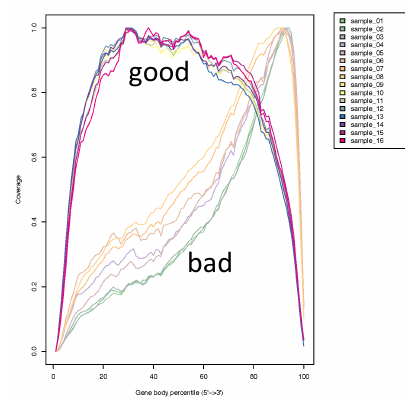
Gene coverage
| * Read coverage of the gene
should be uniform
is expected because of degradation of the RNA
3' bias |
|