Difference between revisions of "Quality Control and Preprocessing Talk"
| Line 45: | Line 45: | ||
= GFF3 file example = | = GFF3 file example = | ||
| − | + | [[File:gff.png]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
= GFF graphicalGFF = | = GFF graphicalGFF = | ||
| − | representation | + | * representation |
| − | structure | + | * structure |
[[File:sascha.png]] | [[File:sascha.png]] | ||
| Line 170: | Line 124: | ||
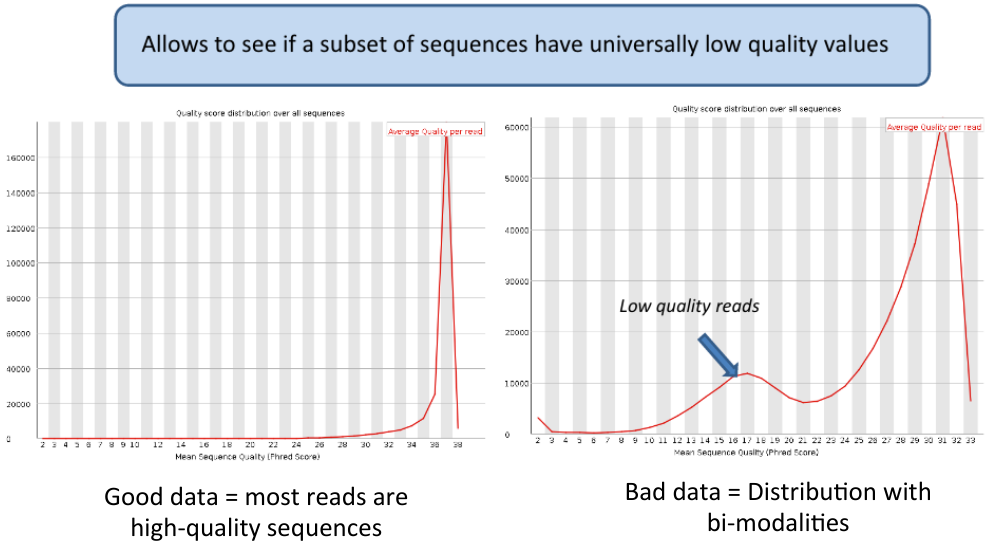
Per sequence quality scores | Per sequence quality scores | ||
| − | [[File: | + | [[File:bimods.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
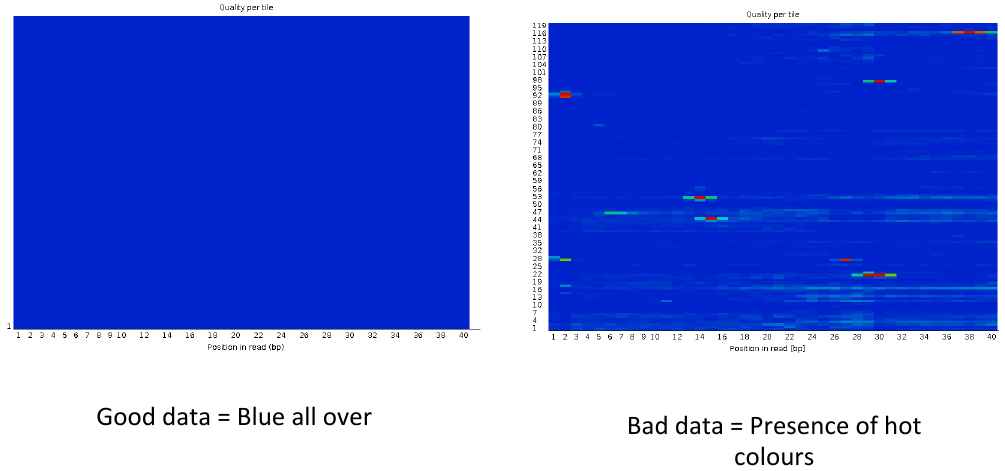
= Per tile sequence quality 1 = | = Per tile sequence quality 1 = | ||
| − | [[File: | + | [[File:ptsq3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
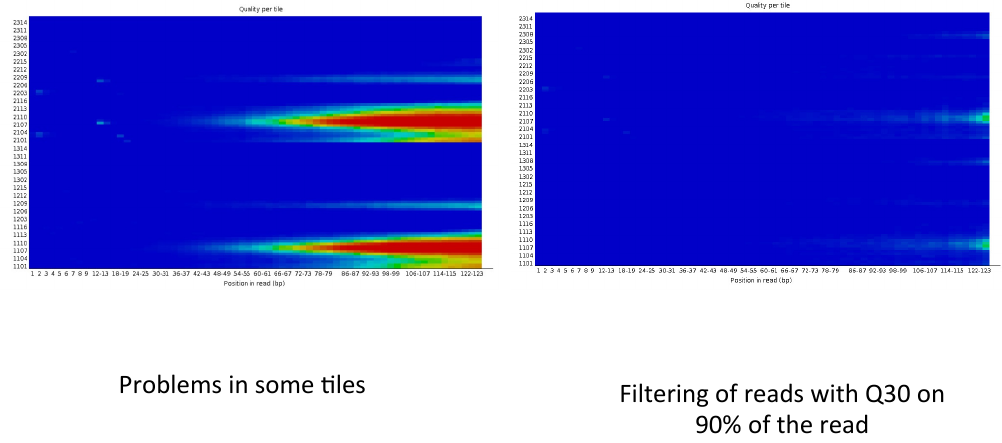
= Per tile sequence quality 2 = | = Per tile sequence quality 2 = | ||
| − | [[File: | + | [[File:ptsq4.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
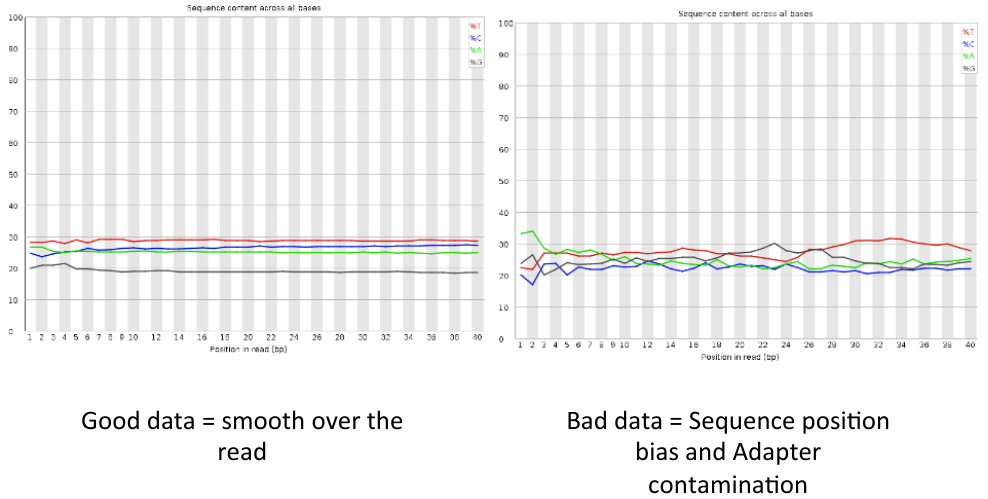
= Per base sequence content = | = Per base sequence content = | ||
| − | [[File: | + | [[File:pbsc3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
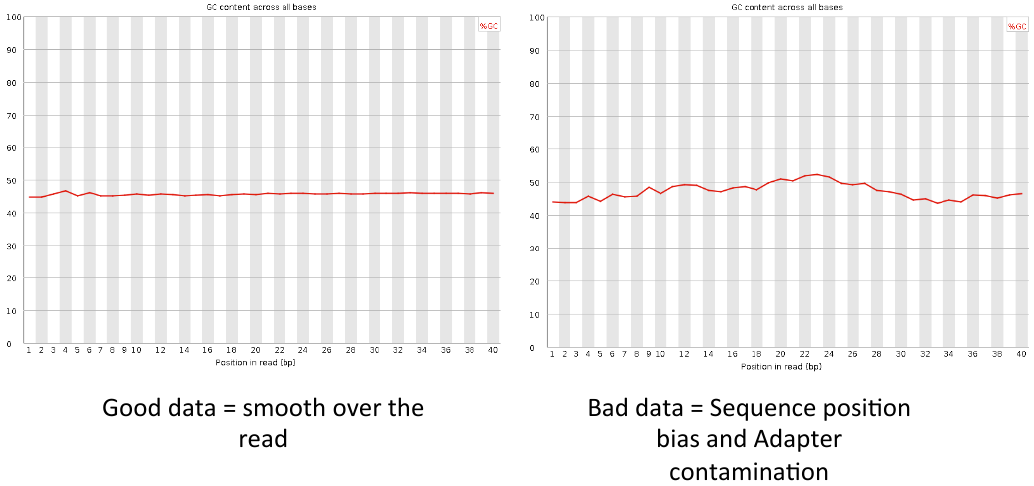
= Per base GC content = | = Per base GC content = | ||
| − | [[File: | + | [[File:pbgcc3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
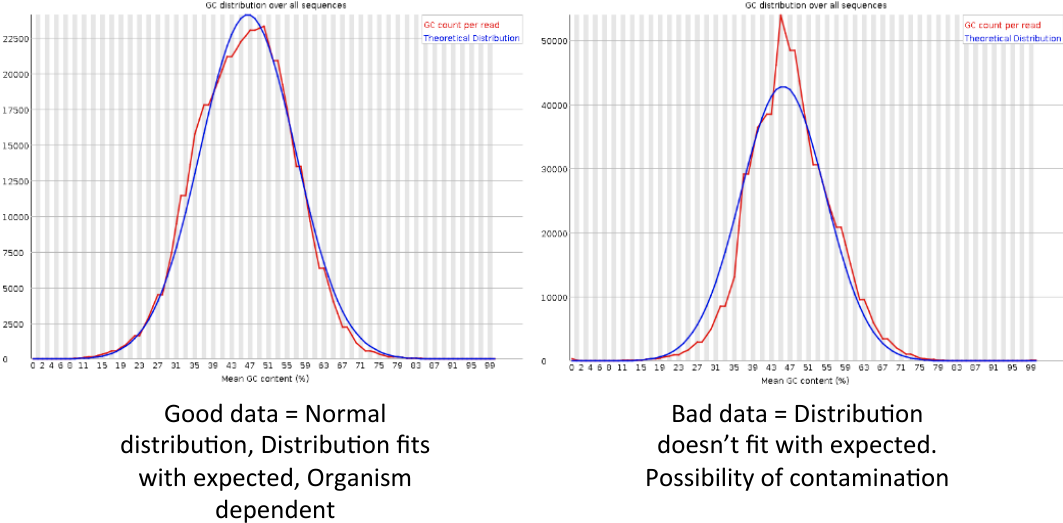
= Per sequence GC content = | = Per sequence GC content = | ||
| − | [[File: | + | [[File:psgcc3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
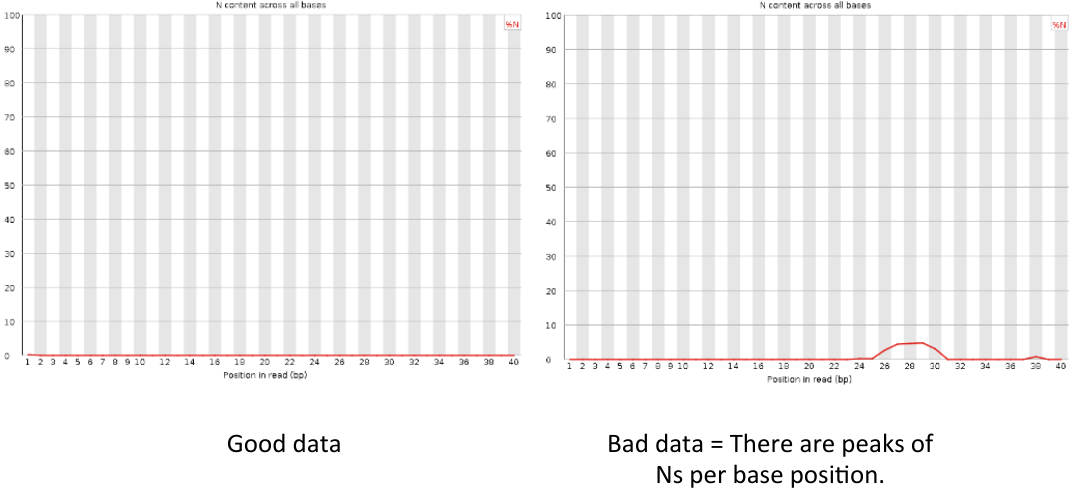
= Per sequence N content = | = Per sequence N content = | ||
| − | [[File: | + | [[File:psnc3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
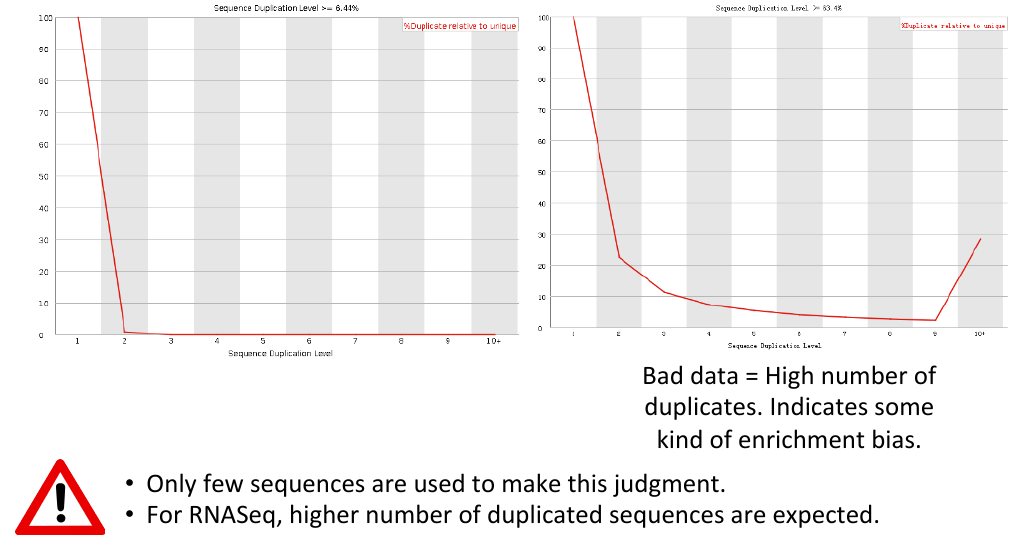
= Sequence duplication levels = | = Sequence duplication levels = | ||
| − | [[File: | + | [[File:sdl3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
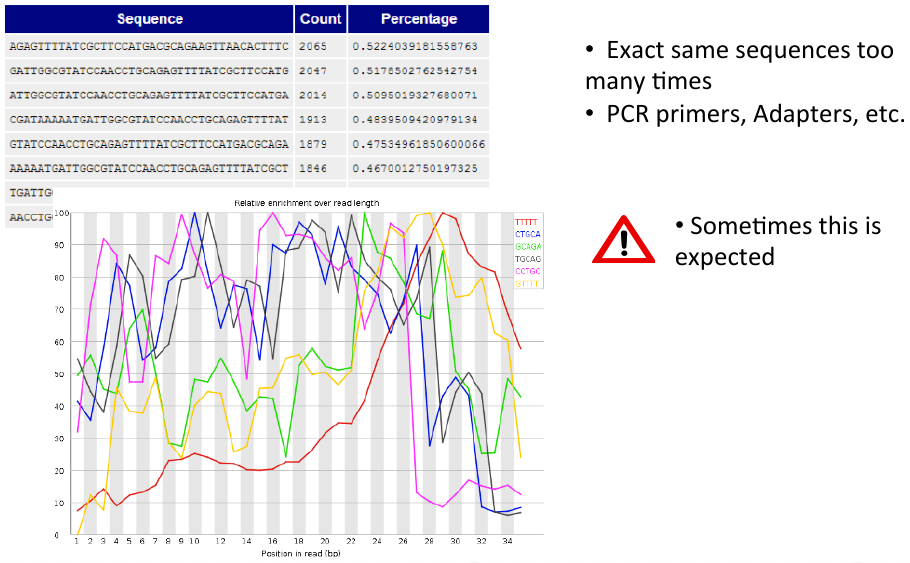
= Overrepresented sequences and k-mer content = | = Overrepresented sequences and k-mer content = | ||
| − | [[File: | + | [[File:ovrep3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
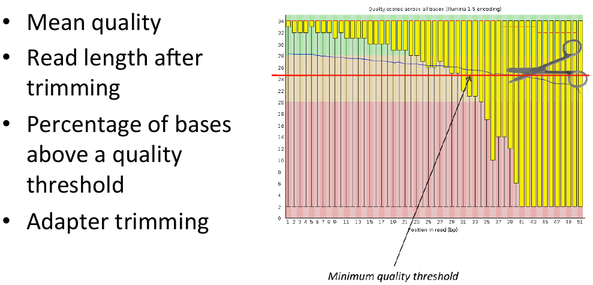
= Sequence Filtering 1 = | = Sequence Filtering 1 = | ||
* It is important to remove bad quality data as our confidence on downstream analysis will be improved. | * It is important to remove bad quality data as our confidence on downstream analysis will be improved. | ||
| − | [[File: | + | [[File:sf1.png]] |
= Sequence Filtering 2 = | = Sequence Filtering 2 = | ||
| − | [[File: | + | [[File:sf3.png]] |
| − | |||
| − | |||
| − | |||
| − | |||
= Sequence filtering tools = | = Sequence filtering tools = | ||
Revision as of 22:02, 8 May 2017
Quality control and data pre-processing
Contents
- 1 Contents
- 2 Data formats
- 3 Data formats
- 4 Sequence Data Format
- 5 GFF
- 6 GFF3 file example
- 7 GFF graphicalGFF
- 8 BED
- 9 Quality Control
- 10 Quality control tools 1
- 11 Quality control tools
- 12 Multiple Sample Quality control
- 13 Addressing QC with FastQC
- 14 Per base sequence quality, good
- 15 Per base sequence quality, bad
- 16 Addressing QC with FastQC
- 17 Per tile sequence quality 1
- 18 Per tile sequence quality 2
- 19 Per base sequence content
- 20 Per base GC content
- 21 Per sequence GC content
- 22 Per sequence N content
- 23 Sequence duplication levels
- 24 Overrepresented sequences and k-mer content
- 25 Sequence Filtering 1
- 26 Sequence Filtering 2
- 27 Sequence filtering tools
- 28 Next
Contents
- Data formats
- – Fasta and Fastq formats
- – Sequence quality encoding
- Quality Control (QC)
- – Evaluation of sequence quality
- – Quality control tools
- – Addressing QC with FastQC
- – Typical artifacts and sequence filtering
Data formats
- Text-based formats
- If not compressed, it can be huge
- Almost every programming language has a parser
Data formats
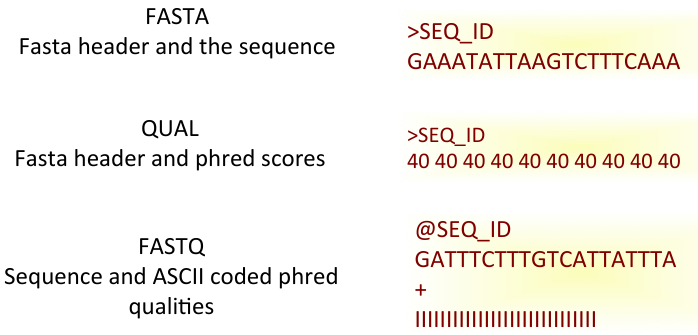
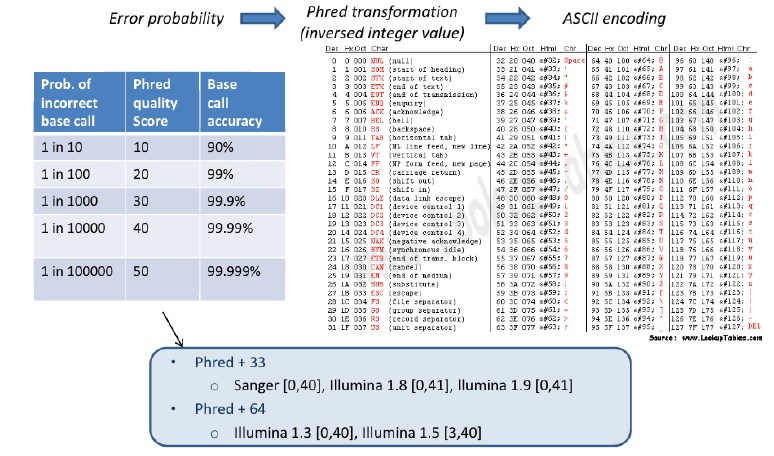
Fastq Format: Sequence quality encoding
Sequence Data Format
Raw sequence data format (Flat/Binary files)
- Fasta, Fastq, HDF5
- Others: http://en.wikipedia.org/wiki/List_of_file_formats#Biology
Processed sequence data format (Flat files)
- Column separated files containing genomic features
and their chromosomal coordinates.
- Different files
- GFF and GTF
- BED
GFF
- Column separated file format contains features located at chromosomal locations
- Not a compact format
- Several versions
– GFF 3 most currently used – GFF 2.5 is also called GTF (used at Ensembl for describing gene features)
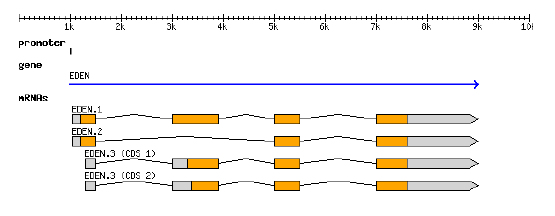
GFF3 file example
GFF graphicalGFF
- representation
- structure
GFF3 can describes the representation of a protein-coding gene (From Sascha Steinbiss' genome-tools suite of programs: http://genometools.org/)
BED
- Created by UCSC Genome team
- Contains similar information to the GFF, but optimized for viewing in the UCSC genome browser
- - Essentially about features and ranges.
- BIG BED, optimized for next gen data – essentially a binary version
- – It can be displayed at UCSC Web browser (even several Gbs !!)
Quality Control
Evaluation of sequence quality
- Primary tool to assess sequencing
- Evaluating sequences in depth is a valuable approach to assess how reliable our results will be.
- QC determines posterior filtering
- - Any filtering decision will affect downstream analysis.
- QC must be run after every critical step.
Quality control tools 1
- Fastx-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html)
- NGS QC Toolkit (http://www.nipgr.res.in/ngsqctoolkit.html)
Quality control tools
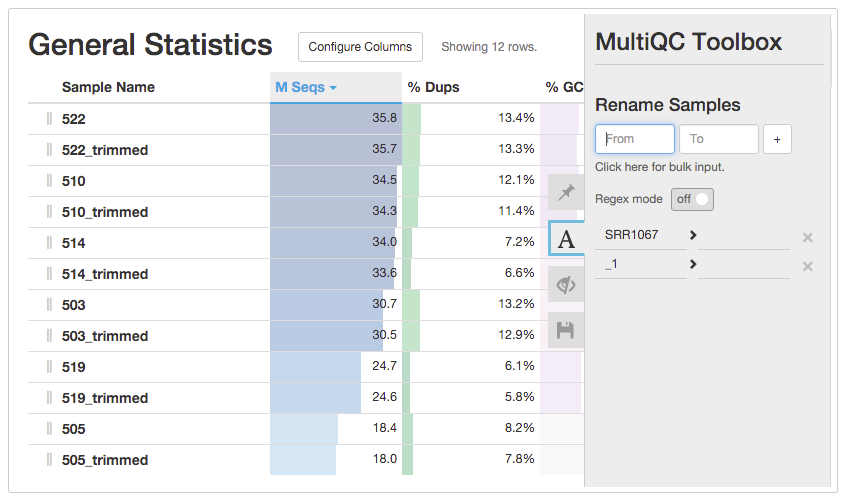
Multiple Sample Quality control
- Multiqc(http://multiqc.info)
- - uses FastQC output
Addressing QC with FastQC
- various screen devoted to plots of the following:
- - Basic stats
- - Per base sequence quality
- - Per read sequence quality
- - Per base sequence content
- - Per base GC content
- - Per sequence GC content
- - Per base N content
- - Sequence length distribution
- - Duplicate sequences
- - Overrepresented sequences
- - Overrepresented k-mers
Examples on web:
- - Good quality:
http://www.bioinformatics.babraham.ac.uk/projects/fastqc/good_sequence_short_fastqc.html
- - Bad quality:
http://www.bioinformatics.babraham.ac.uk/projects/fastqc/bad_sequence_fastqc.html
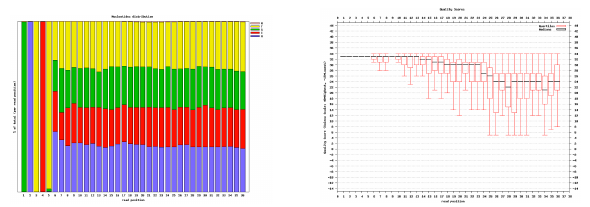
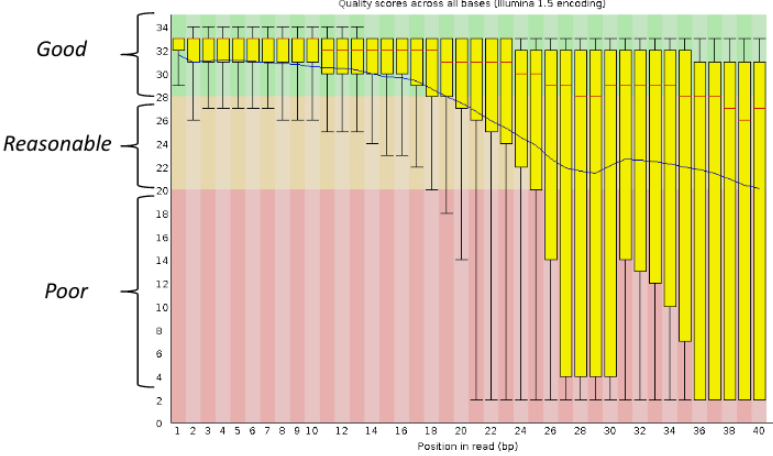
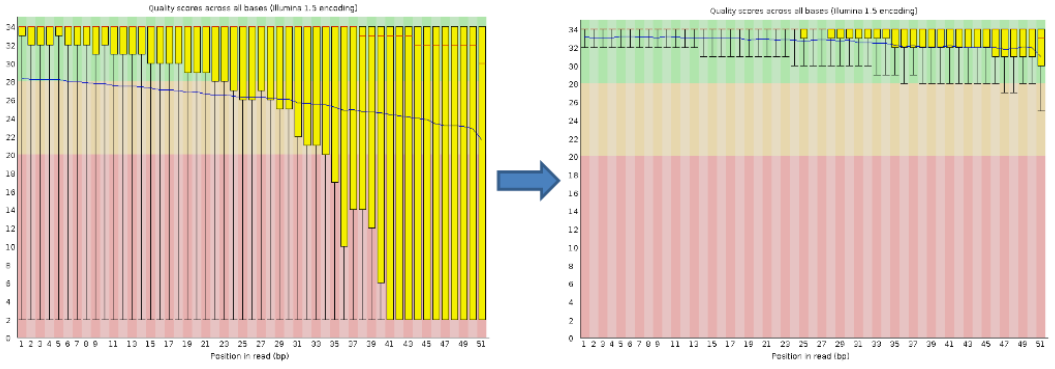
Per base sequence quality, good
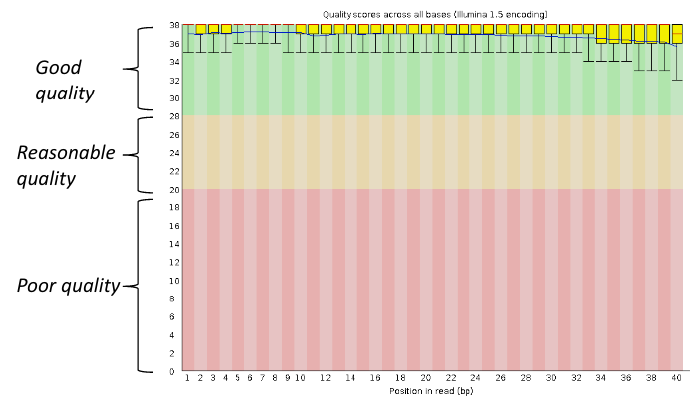 Good data = Consistent high quality along the read
Good data = Consistent high quality along the read
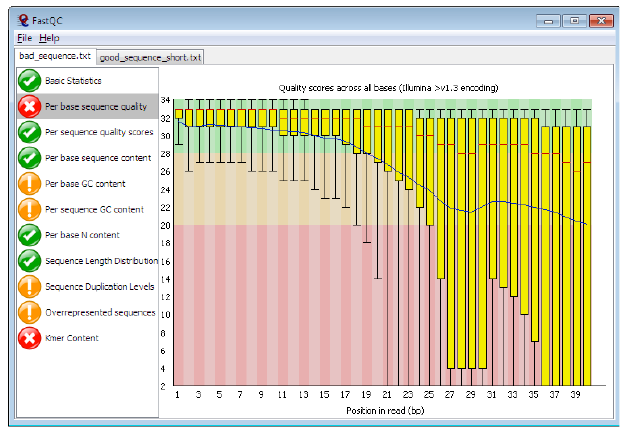
Per base sequence quality, bad
Bad data = Quality decreases towards the end of the read and High variance
Addressing QC with FastQC
Per sequence quality scores
Per tile sequence quality 1
Per tile sequence quality 2
Per base sequence content
Per base GC content
Per sequence GC content
Per sequence N content
Sequence duplication levels
Overrepresented sequences and k-mer content
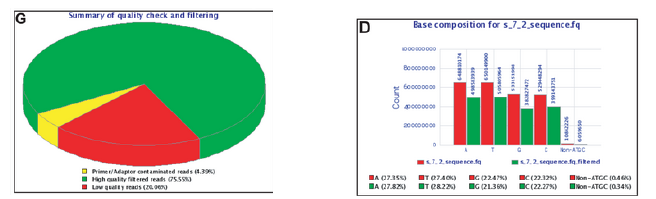
Sequence Filtering 1
- It is important to remove bad quality data as our confidence on downstream analysis will be improved.
Sequence Filtering 2
Sequence filtering tools
- Fastq-mcf
- Cutadapt
- SeqTK
- Trimmomatic
Next
Practical sequence filtering session