Difference between revisions of "ChIP-Seq Top2 peak-calling"
m (Protected "ChIP-Seq Top2 peak-calling" ([Edit=Allow only administrators] (indefinite) [Move=Allow only administrators] (indefinite))) |
|
(No difference)
| |
Latest revision as of 09:19, 4 June 2018
Contents
Introduction
ChIP-Seq on Mutant vs. Wildtype, two states, 4 sets of pairs each, for a total of 16 pairs of paired-end datasets.
The generic pipeline
One of the authors also has a tutorial which is formatted into wiki format (for easy copy-pasting) here.
Exp 1: Browsing alignment to S288C reference genome
This is the link to the genome browser.
- Tick items in the left hand panel to enable/disable visibility
- Hover mouse over loci position bar, it will change into a plus sign. Drag a region to zoom quickly into it.
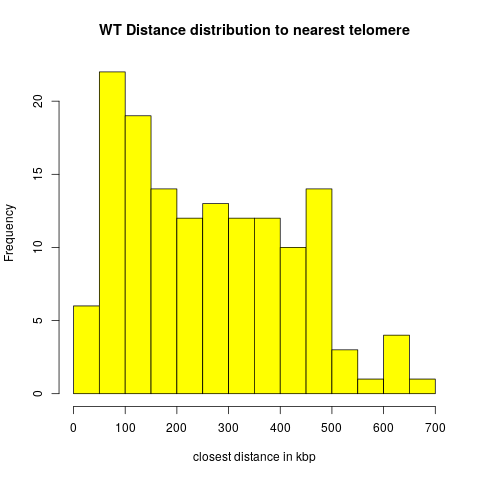
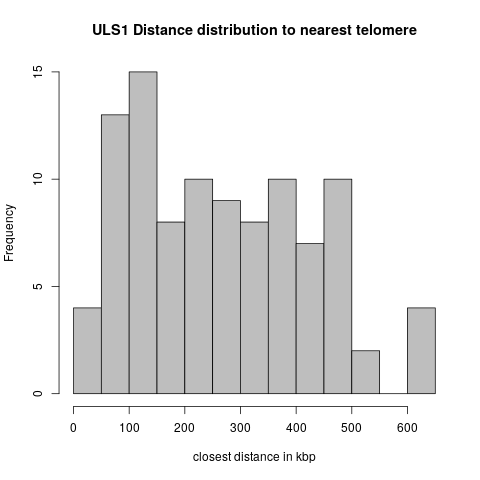
Exp1: Distances to Telomeres
- Of the 166 wildtype peak regions, 22 overlaps with telomeres.
- Of the 124 ULS1 peak regions, 23 overlap with telomeres.
These cases are excluded from the distance distributions below.
Wildtype
Uls1
Raw data description
Mutant:
ULS1INP_S6_L001_R1_001.fastq.gz ULS1INP_S6_L001_R2_001.fastq.gz ULS1INP_S6_L002_R1_001.fastq.gz ULS1INP_S6_L002_R2_001.fastq.gz ULS1INP_S6_L003_R1_001.fastq.gz ULS1INP_S6_L003_R2_001.fastq.gz ULS1INP_S6_L004_R1_001.fastq.gz ULS1INP_S6_L004_R2_001.fastq.gz USL1IP_S7_L001_R1_001.fastq.gz USL1IP_S7_L001_R2_001.fastq.gz USL1IP_S7_L002_R1_001.fastq.gz USL1IP_S7_L002_R2_001.fastq.gz USL1IP_S7_L003_R1_001.fastq.gz USL1IP_S7_L003_R2_001.fastq.gz USL1IP_S7_L004_R1_001.fastq.gz USL1IP_S7_L004_R2_001.fastq.gz
Wildtype:
WTINP_S4_L001_R1_001.fastq.gz WTINP_S4_L001_R2_001.fastq.gz WTINP_S4_L002_R1_001.fastq.gz WTINP_S4_L002_R2_001.fastq.gz WTINP_S4_L003_R1_001.fastq.gz WTINP_S4_L003_R2_001.fastq.gz WTINP_S4_L004_R1_001.fastq.gz WTINP_S4_L004_R2_001.fastq.gz WTIP_S5_L001_R1_001.fastq.gz WTIP_S5_L001_R2_001.fastq.gz WTIP_S5_L002_R1_001.fastq.gz WTIP_S5_L002_R2_001.fastq.gz WTIP_S5_L003_R1_001.fastq.gz WTIP_S5_L003_R2_001.fastq.gz WTIP_S5_L004_R1_001.fastq.gz WTIP_S5_L004_R2_001.fastq.gz
Raw dataset quality analysis
Using FASTQC and them MultiQC.
Results linked here.
Adaptor-cutting and filtering procedure
- First run was without any filtering or cutting.
BAM file quality analysis
Using BamQC.
For the unfiltered reads
Results linked here.
Papers
Genome-Organizing Factors Top2 and Hmo1 Prevent Chromosome Fragility at Sites of S phase Transcription
link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16258
Abstract:
Specialized topoisomerases solve the topological constraints arising when replication forks encounter transcription. We have investigated the contribution of Top2 in S phase transcription. Specifically in S phase, Top2 binds intergenic regions close to transcribed genes. The Top2-bound loci exhibit low nucleosome density and accumulate gammaH2A when Top2 is defective. These intergenic loci associate with the HMG protein Hmo1 throughout the cell cycle and are refractory to the histone variant Htz1. In top2 mutants, Hmo1 is deleterious and accumulates at pericentromeric regions in G2/M. Our data indicate that Top2 is dispensable for transcription and that Hmo1 and Top2 bind in the proximity of genes transcribed in S phase suppressing chromosome fragility at the M-G1 transition. We propose that an Hmo1-dependent epigenetic signature together with Top2 mediate an S phase architectural pathway to preserve genome integrity.
Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast
link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22626
DNA topoisomerases are believed to promote transcription by removing excessive DNA supercoils produced during elongation. However, it is unclear how topoisomerases in eukaryotes are recruited and function in the transcription pathway in the context of nucleosomes. To address this problem we present high-resolution genome-wide maps of one of the major eukaryotic topoisomerases, Topoisomerase II (Top2) and nucleosomes in the budding yeast, Saccharomyces cerevisiae. Our data indicate that at promoters Top2 binds primarily to DNA that is nucleosome-free. However, although nucleosome loss enables Top2 occupancy, the opposite is not the case and the loss of Top2 has little effect on nucleosome density. We also find that Top2 is involved in transcription. Not only is Top2 enriched at highly transcribed genes, but Top2 is required redundantly with Top1 for optimal recruitment of RNA polymerase II at their promoters. These findings and the examination of candidate-activated genes suggest that nucleosome loss induced by nucleosome remodeling factors during gene activation enables Top2 binding, which in turn acts redundantly with Top1 to enhance recruitment of RNA polymerase II.