Difference between revisions of "CallSNPs.py"
| (103 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | = Introduction = | |
| − | + | The call SNPS.py script is part of the '''script_tools''' environment-module, and is loaded with: | |
| − | + | module load script_tools | |
| − | + | As input, it requires an alignment in the shape of the base reference sequence and the bam files (prefereably sorted) of the short reads aligned to that reference. So clearly, that step should have already been taken before using CallSNPs.py. The recommended tools for that are '''bwa''' and '''bowtie'''. | |
| − | |||
| − | + | The output of callSNPs.py is a fasta file, because its last step is from vcf-tools package: | |
| + | vcf_tab_to_fasta_alignment.pl | ||
| − | + | = Pipeline Summary = | |
| − | |||
| − | |||
| − | 3. | + | {|class="wikitable" style="width:90%" |
| − | + | |style="width: 3%; text-align:right;"|'''Step''' | |
| + | |style="width: 14%"|'''Tool''' | ||
| + | |style="width: 4%"|'''Version''' | ||
| + | |style="width: 33%"|'''Operation''' | ||
| + | |style="width: 25%"|'''Inputs''' | ||
| + | |style="width: 20%"|'''Outputs''' | ||
| + | |- | ||
| + | |style="text-align:right;"|'''1.''' | ||
| + | |bwa | ||
| + | |0.7.15 | ||
| + | |Alignment/Mapping of reads to reference | ||
| + | |Reference FASTA and FASTQ reads | ||
| + | |BAM files | ||
| + | |- | ||
| + | |style="text-align:right;"|'''2.''' | ||
| + | |samtools mpileup | ||
| + | |0.1.19 | ||
| + | |Calculation of SNP values | ||
| + | |BAM files | ||
| + | |BCF files | ||
| + | |- | ||
| + | |style="text-align:right;"|'''3.''' | ||
| + | |bcftools | ||
| + | |0.1.19 | ||
| + | |Classification into SNPs | ||
| + | |BCF files | ||
| + | |VCF files | ||
| + | |- | ||
| + | |style="text-align:right;"|'''4.''' | ||
| + | |bedtools genomecov | ||
| + | |Gitv6114307 | ||
| + | |Generate high coverage region file | ||
| + | |BAM files | ||
| + | |BED files | ||
| + | |- | ||
| + | |style="text-align:right;"|'''5.''' | ||
| + | |bedtools intersect | ||
| + | |Gitv6114307 | ||
| + | |Find regions common to whole dataset | ||
| + | |all BED files | ||
| + | |single intersect BED file | ||
| + | |- | ||
| + | |style="text-align:right;"|'''6.''' | ||
| + | |vcftools | ||
| + | |Gitv4fce995 | ||
| + | |Recode VCF files based on intersect regions | ||
| + | |single intersect BED file | ||
| + | |Recoded VCF files | ||
| + | |- | ||
| + | |style="text-align:right;"|'''7.''' | ||
| + | |bcftools filter | ||
| + | |1.2 | ||
| + | |Filter out <30 base and <50 mapping quality | ||
| + | |BCF files | ||
| + | |VCF files | ||
| + | |- | ||
| + | |style="text-align:right;"|'''9.''' | ||
| + | |vcftools vcf-merge | ||
| + | |Gitv4fce995 | ||
| + | |Merge all VCF file into one | ||
| + | |Multiple recoded VCF files | ||
| + | |Single VCF file | ||
| + | |- | ||
| + | |style="text-align:right;"|'''9.''' | ||
| + | |vcftools | ||
| + | |Gitv4fce995 | ||
| + | |Convert merged VCF file to sequences | ||
| + | |Single VCF files | ||
| + | |Single FASTA file | ||
| + | |- | ||
| + | |style="text-align:right;"|'''10.''' | ||
| + | |RAxML | ||
| + | |Gitv4692545 | ||
| + | |Calculate tree using Ascertainment Bias | ||
| + | |Single FASTA file | ||
| + | |Newick file | ||
| + | |} | ||
| − | '''bedtools genomecov''' outputs the number of aligned short-read bases in the genome in a format (enabled by the '''-bga''' option) in the fourth column. This is picked up by the '''awk''' which ignores all those less than '''NUM'''. If '''NUM''' is set to zero, the entire variant callset will be returned in raw form | + | Note: The first and final steps, 1 and 10 are not automated into the '''CallSNPS.py''' pipeline and must be carried out manually. |
| + | |||
| + | = How to run = | ||
| + | |||
| + | Here is an example jobscript to run on the cluster | ||
| + | |||
| + | #!/bin/bash | ||
| + | #$ -cwd | ||
| + | #$ -j y | ||
| + | #$ -S /bin/bash | ||
| + | #$ -V | ||
| + | #$ -q marvin.q | ||
| + | callSNPs.py -t name -i altsortedbam/*.bam -r contigs.fa -o out_report_alt.txt | ||
| + | |||
| + | |||
| + | = Details of pipeline stages = | ||
| + | |||
| + | == Layout == | ||
| + | |||
| + | The following images summarizes the pipeline. A detailed explanation then follows: | ||
| + | |||
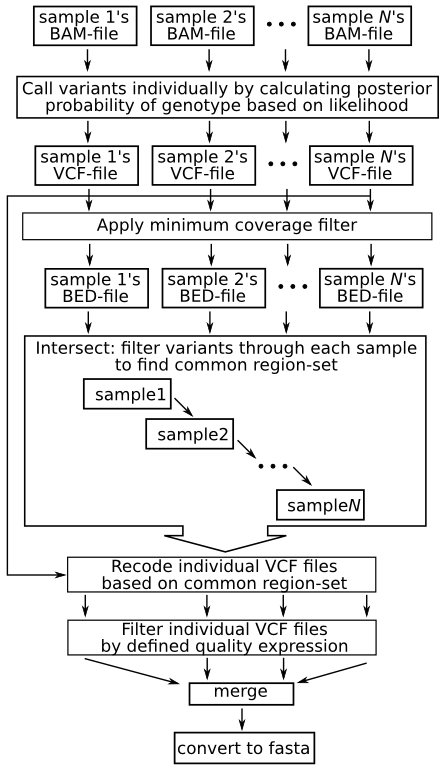
| + | [[File:Callsnppipe.png]] | ||
| + | |||
| + | == Description == | ||
| + | |||
| + | ===Calculate likelihoods of each possible variant, and apply prior as well as call variants=== | ||
| + | samtools mpileup -E -d ''NUM'' -DSug -Q 1 -f ''ref_file'' ''input_bam_file'' | bcftools view -u - > ''output_bcf_file'' | ||
| + | |||
| + | <ins>Explanation</ins>: | ||
| + | * '''mpileup''' collects summary information from the input BAMs and then computes and stores the likelihood of data given each possible genotype. | ||
| + | * '''-E''', recalculates BAQ (Base Alignment Quality) as well. The existing BQ tags ae ignored (these are probably previous BAQ values). | ||
| + | * '''-d''', max depth value. From manual "at a position, read maximally INT reads per input file". In this script the value of 1500 is the default. | ||
| + | * '''-D''', output per sample read depth. The new style is -t DP which explicitly matches with VCF file header tags. | ||
| + | * '''-S''', phred-scaled strand bias p-value. The new style is -t SP, above rationale. | ||
| + | * '''-u''', ensures output is uncompressed, which is better for the piping that takes palce here. | ||
| + | * '''-g''', output in BCF | ||
| + | * '''-B''', disable realignment based on BAQ. Particularly avoids false SNP causing by malalignment which is critical in this pipeline | ||
| + | * '''-Q''', minimum BQ, presumably this is based on the recalculated BQ from the '''-E''' option above. | ||
| + | * '''-f''', this is for referencedicates the name of the reference file is next. | ||
| + | * '''bcftools view''' applies prior and then calls the variants. Can be quite raw so often a filter is needed after this, which will be the next step. | ||
| + | * '''-g''', print sites with at least one genotype | ||
| + | * '''-u''', print sites with uncalled genotypes (with the '''-g''' options this means almost all sites, one presumes) | ||
| + | |||
| + | <ins>Comments on original</ins>: | ||
| + | The original samtools mpileup command had an '''-E''' option at the beginning of the option list. But this is incompatible with the '''-B''' which is more important, so it was removed. | ||
| + | * '''-E''', recalculates BAQ (Base Alignment Quality) as well. The existing BQ tags ae ignored (these are probably previous BAQ values). | ||
| + | The original bcftools view part included a '''-c''', but: | ||
| + | * '''-c''', miniimum allele count. Would require a value but not here. | ||
| + | * '''-b''', unidentified option. bcftools man page does not register this for either in common bcftools commands nor specifically in '''bcftools view'''. | ||
| + | There was also a difficulty with the '''-g''' option: | ||
| + | The argument to -g not recognised. Expected one of hom/het/miss/^hom/^het/^miss, got "u". | ||
| + | So it was also left out. | ||
| + | |||
| + | ===Filter variants through a maximum read depth === | ||
| + | bcftools view ''bcf_file'' | vcfutils.pl varFilter -D''NUM'' > ''outputvcffilename'' | ||
| + | '''bcftools view''' outputs the variants in VCF format and '''vcfutils.pl''' perl script , which is part of samtools platform, filters for variants on their maximum value via the '''-D''' option (the default for this is 10 million, which strikes one as meaning "infinite"). A more normal value is 1500 (and this is the default value in the original script). | ||
| + | |||
| + | ===Housekeeping: compress vcf files=== | ||
| + | Not strictly part of the sequential run of the pipeline, but at this point the vcf files are compressed with '''samtools'''' bgzip and then indexed with '''tabix''' (also from '''samtools''', a generic tab-delimited file indexer). | ||
| + | bgzip -c ''vcf_file'' > ''vcf_file.gz'' | ||
| + | tabix -p vcf ''vcf_file.gz'' | ||
| + | The latter command creates an extra ''vcf_file.gz.tbi'' file in the directory. | ||
| + | |||
| + | ===Extract features with minimum coverage === | ||
| + | bedtools genomecov -bga -ibam ''input_bam_file'' | awk -v t=''NUM'' "{if($4>=t)print}" | bedtools merge -i - > ''output_bed_file'' | ||
| + | |||
| + | '''bedtools genomecov''' outputs the number of aligned short-read bases in the genome in a format (enabled by the '''-bga''' option) in the fourth column. This is picked up by the '''awk''' program which ignores all those less than '''NUM'''. If '''NUM''' is set to zero, the entire variant callset will be returned in raw form because nothing will be filtered and the '''-bga''' (with '''-a''' presumably standing for ''all'') will include areas with zero coverage (this sounds uninteresting but it could be useful to have, so it's often kept). The default value for ''NUM'' set in the overall python script is 15. The final merge, as well as merging the filtered output, also makes sure that split and overlapping features are rolled into one. Note that at this point our variant format VCF turns into a BED file, and we move from talking about variants to talking about regions or features, which is what bed files record. | ||
| + | |||
| + | ===Intersect samples' features to find common feature-set === | ||
| + | With every short read alignment (i.e. ''BAM'' file) now having its set of features in a corresponding ''BED'' file, we look to find the features that all samples share by using '''bedtools'''' intersect subcommand. So for each sample's ''BED'' file, the following is executed> | ||
| + | bedtools intersect -a ''cumulative_intersect_bed_file'' -b ''sample_bed_file'' > ''single_intersect_bed_file'' | ||
| + | mv ''single_intersect_file'' ''cumulative_intersect_file'' | ||
| + | |||
| + | At the end, the cumulative intersect file is renamed so we can know its work is finished. | ||
| + | mv ''cumulative_intersect_bed_file'' ''final_intersect_bed_file'' | ||
| + | |||
| + | ===Recode vcf files to reflect common feature set=== | ||
| + | We now want to recode our sample vcf files taking into account the common regions now described in the intersect feature set captured in the '''''cumulative_intersection_file''''' above so that the common variants can be excluded, because we are not interested in the similarities of the samples, but rather the differences. | ||
| + | vcftools --gzvcf ''sample_vcf_file'' --bed ''final_intersect_bed_file'' --out ''recode_file_prefix'' --recode --keep-INFO-all | ||
| + | This can all be performed on the compressed vcf files. The '''recode.vcf''' will be appended to the prefix specified above. | ||
| + | |||
| + | ===Filter recoded VCF file by quality specification === | ||
| + | |||
| + | There are certain quality consideration we want the VCF files to take into account, which '''samtools'''' ''bcftools filter'' command can achieve for us. The quality specification or expression involves merging different aspects of its value via the logical OR operation, in the shapes of i double-bar: ''||'': | ||
| + | |||
| + | VCF_QUALITY_EXPRESSION = '%TYPE="indel" || %TYPE="mnp" || %TYPE="other" || %QUAL<50 || INFO/DP<4 || INFO/DP4[3]<2 || INFO/DP4[4]<2 || INFO/MQ<30 || INFO/AF1<0.95 || INFO/PV4[0]<0.001 || INFO/PV4[2]<0.001 || INFO/PV4[3]<0.001' | ||
| + | |||
| + | This can be inserted into the ''bcftools''' command-line as follows: | ||
| + | |||
| + | bcftools filter -e ''VCF_QUALITY_EXPRESSION'' ''sample_vcf_file'' | bgzip -c > ''compressed_sample_vcf_file'' | ||
| + | |||
| + | Note how the vcf output is also compressed and can also be indexed: | ||
| + | |||
| + | tabix ''compressed_sample_vcf_file'' | ||
| + | |||
| + | ===Merge VCF files === | ||
| + | The ''vcftools'' program suite has a simple command for merging compressed vcfs, although its output is uncompressed, so we need to compress it also via a unix pipe (the bar ''|'') | ||
| + | vcf-merge ''all_compressed_vcf_files'' | bgzip -c > ''merged_vcf_file_compressed'' | ||
| + | |||
| + | ===Create fasta file from merged VCF file === | ||
| + | |||
| + | First however we need to render the VCF file into tabbed format, for which ''vcftools'' has the ''vcf-to-tab'' subcommand: | ||
| + | zcat ''merged_vcf_file_compressed'' | vcf-to-tab > ''merged_tabbed_vcf_file'' | ||
| + | |||
| + | To convert to fasta format, there is a perl script: | ||
| + | |||
| + | vcf_tab_to_fasta_alignment.pl --exclude_het -i ''merged_tabbed_vcf_file'' > ''merged_common_variants_fasta'' | ||
| + | |||
| + | The ID lines of the resulting fasta file needs to be cleaned a little to ensure later downstream compatibility. | ||
| + | |||
| + | = Links = | ||
| + | * A similar pipeline from [https://www.ebi.ac.uk/sites/ebi.ac.uk/files/content.ebi.ac.uk/materials/2014/140217_AgriOmics/dan_bolser_snp_calling.pdf Dan Bolser]. | ||
Latest revision as of 09:27, 6 October 2017
Contents
- 1 Introduction
- 2 Pipeline Summary
- 3 How to run
- 4 Details of pipeline stages
- 4.1 Layout
- 4.2 Description
- 4.2.1 Calculate likelihoods of each possible variant, and apply prior as well as call variants
- 4.2.2 Filter variants through a maximum read depth
- 4.2.3 Housekeeping: compress vcf files
- 4.2.4 Extract features with minimum coverage
- 4.2.5 Intersect samples' features to find common feature-set
- 4.2.6 Recode vcf files to reflect common feature set
- 4.2.7 Filter recoded VCF file by quality specification
- 4.2.8 Merge VCF files
- 4.2.9 Create fasta file from merged VCF file
- 5 Links
Introduction
The call SNPS.py script is part of the script_tools environment-module, and is loaded with:
module load script_tools
As input, it requires an alignment in the shape of the base reference sequence and the bam files (prefereably sorted) of the short reads aligned to that reference. So clearly, that step should have already been taken before using CallSNPs.py. The recommended tools for that are bwa and bowtie.
The output of callSNPs.py is a fasta file, because its last step is from vcf-tools package:
vcf_tab_to_fasta_alignment.pl
Pipeline Summary
| Step | Tool | Version | Operation | Inputs | Outputs |
| 1. | bwa | 0.7.15 | Alignment/Mapping of reads to reference | Reference FASTA and FASTQ reads | BAM files |
| 2. | samtools mpileup | 0.1.19 | Calculation of SNP values | BAM files | BCF files |
| 3. | bcftools | 0.1.19 | Classification into SNPs | BCF files | VCF files |
| 4. | bedtools genomecov | Gitv6114307 | Generate high coverage region file | BAM files | BED files |
| 5. | bedtools intersect | Gitv6114307 | Find regions common to whole dataset | all BED files | single intersect BED file |
| 6. | vcftools | Gitv4fce995 | Recode VCF files based on intersect regions | single intersect BED file | Recoded VCF files |
| 7. | bcftools filter | 1.2 | Filter out <30 base and <50 mapping quality | BCF files | VCF files |
| 9. | vcftools vcf-merge | Gitv4fce995 | Merge all VCF file into one | Multiple recoded VCF files | Single VCF file |
| 9. | vcftools | Gitv4fce995 | Convert merged VCF file to sequences | Single VCF files | Single FASTA file |
| 10. | RAxML | Gitv4692545 | Calculate tree using Ascertainment Bias | Single FASTA file | Newick file |
Note: The first and final steps, 1 and 10 are not automated into the CallSNPS.py pipeline and must be carried out manually.
How to run
Here is an example jobscript to run on the cluster
#!/bin/bash #$ -cwd #$ -j y #$ -S /bin/bash #$ -V #$ -q marvin.q callSNPs.py -t name -i altsortedbam/*.bam -r contigs.fa -o out_report_alt.txt
Details of pipeline stages
Layout
The following images summarizes the pipeline. A detailed explanation then follows:
Description
Calculate likelihoods of each possible variant, and apply prior as well as call variants
samtools mpileup -E -d NUM -DSug -Q 1 -f ref_file input_bam_file | bcftools view -u - > output_bcf_file
Explanation:
- mpileup collects summary information from the input BAMs and then computes and stores the likelihood of data given each possible genotype.
- -E, recalculates BAQ (Base Alignment Quality) as well. The existing BQ tags ae ignored (these are probably previous BAQ values).
- -d, max depth value. From manual "at a position, read maximally INT reads per input file". In this script the value of 1500 is the default.
- -D, output per sample read depth. The new style is -t DP which explicitly matches with VCF file header tags.
- -S, phred-scaled strand bias p-value. The new style is -t SP, above rationale.
- -u, ensures output is uncompressed, which is better for the piping that takes palce here.
- -g, output in BCF
- -B, disable realignment based on BAQ. Particularly avoids false SNP causing by malalignment which is critical in this pipeline
- -Q, minimum BQ, presumably this is based on the recalculated BQ from the -E option above.
- -f, this is for referencedicates the name of the reference file is next.
- bcftools view applies prior and then calls the variants. Can be quite raw so often a filter is needed after this, which will be the next step.
- -g, print sites with at least one genotype
- -u, print sites with uncalled genotypes (with the -g options this means almost all sites, one presumes)
Comments on original: The original samtools mpileup command had an -E option at the beginning of the option list. But this is incompatible with the -B which is more important, so it was removed.
- -E, recalculates BAQ (Base Alignment Quality) as well. The existing BQ tags ae ignored (these are probably previous BAQ values).
The original bcftools view part included a -c, but:
- -c, miniimum allele count. Would require a value but not here.
- -b, unidentified option. bcftools man page does not register this for either in common bcftools commands nor specifically in bcftools view.
There was also a difficulty with the -g option:
The argument to -g not recognised. Expected one of hom/het/miss/^hom/^het/^miss, got "u".
So it was also left out.
Filter variants through a maximum read depth
bcftools view bcf_file | vcfutils.pl varFilter -DNUM > outputvcffilename
bcftools view outputs the variants in VCF format and vcfutils.pl perl script , which is part of samtools platform, filters for variants on their maximum value via the -D option (the default for this is 10 million, which strikes one as meaning "infinite"). A more normal value is 1500 (and this is the default value in the original script).
Housekeeping: compress vcf files
Not strictly part of the sequential run of the pipeline, but at this point the vcf files are compressed with samtools' bgzip and then indexed with tabix (also from samtools, a generic tab-delimited file indexer).
bgzip -c vcf_file > vcf_file.gz tabix -p vcf vcf_file.gz
The latter command creates an extra vcf_file.gz.tbi file in the directory.
Extract features with minimum coverage
bedtools genomecov -bga -ibam input_bam_file | awk -v t=NUM "{if($4>=t)print}" | bedtools merge -i - > output_bed_file
bedtools genomecov outputs the number of aligned short-read bases in the genome in a format (enabled by the -bga option) in the fourth column. This is picked up by the awk program which ignores all those less than NUM. If NUM is set to zero, the entire variant callset will be returned in raw form because nothing will be filtered and the -bga (with -a presumably standing for all) will include areas with zero coverage (this sounds uninteresting but it could be useful to have, so it's often kept). The default value for NUM set in the overall python script is 15. The final merge, as well as merging the filtered output, also makes sure that split and overlapping features are rolled into one. Note that at this point our variant format VCF turns into a BED file, and we move from talking about variants to talking about regions or features, which is what bed files record.
Intersect samples' features to find common feature-set
With every short read alignment (i.e. BAM file) now having its set of features in a corresponding BED file, we look to find the features that all samples share by using bedtools' intersect subcommand. So for each sample's BED file, the following is executed>
bedtools intersect -a cumulative_intersect_bed_file -b sample_bed_file > single_intersect_bed_file mv single_intersect_file cumulative_intersect_file
At the end, the cumulative intersect file is renamed so we can know its work is finished.
mv cumulative_intersect_bed_file final_intersect_bed_file
Recode vcf files to reflect common feature set
We now want to recode our sample vcf files taking into account the common regions now described in the intersect feature set captured in the cumulative_intersection_file above so that the common variants can be excluded, because we are not interested in the similarities of the samples, but rather the differences.
vcftools --gzvcf sample_vcf_file --bed final_intersect_bed_file --out recode_file_prefix --recode --keep-INFO-all
This can all be performed on the compressed vcf files. The recode.vcf will be appended to the prefix specified above.
Filter recoded VCF file by quality specification
There are certain quality consideration we want the VCF files to take into account, which samtools' bcftools filter command can achieve for us. The quality specification or expression involves merging different aspects of its value via the logical OR operation, in the shapes of i double-bar: ||:
VCF_QUALITY_EXPRESSION = '%TYPE="indel" || %TYPE="mnp" || %TYPE="other" || %QUAL<50 || INFO/DP<4 || INFO/DP4[3]<2 || INFO/DP4[4]<2 || INFO/MQ<30 || INFO/AF1<0.95 || INFO/PV4[0]<0.001 || INFO/PV4[2]<0.001 || INFO/PV4[3]<0.001'
This can be inserted into the bcftools' command-line as follows:
bcftools filter -e VCF_QUALITY_EXPRESSION sample_vcf_file | bgzip -c > compressed_sample_vcf_file
Note how the vcf output is also compressed and can also be indexed:
tabix compressed_sample_vcf_file
Merge VCF files
The vcftools program suite has a simple command for merging compressed vcfs, although its output is uncompressed, so we need to compress it also via a unix pipe (the bar |)
vcf-merge all_compressed_vcf_files | bgzip -c > merged_vcf_file_compressed
Create fasta file from merged VCF file
First however we need to render the VCF file into tabbed format, for which vcftools has the vcf-to-tab subcommand:
zcat merged_vcf_file_compressed | vcf-to-tab > merged_tabbed_vcf_file
To convert to fasta format, there is a perl script:
vcf_tab_to_fasta_alignment.pl --exclude_het -i merged_tabbed_vcf_file > merged_common_variants_fasta
The ID lines of the resulting fasta file needs to be cleaned a little to ensure later downstream compatibility.
Links
- A similar pipeline from Dan Bolser.