Difference between revisions of "Intro to RNA-Seq Data Analysis Course"
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
= Course schedule = | = Course schedule = | ||
| − | * This is based on a 2 day Edinburgh Genomics course of the same name | + | * This is based on a 2 day Edinburgh Genomics course of the same name, with the following changes: |
| − | + | :- "Introduction to Linux" moduel excluded | |
| − | + | :- "Sequencer technology overview" module excluded. | |
| − | + | :- No laboratory visit | |
| + | :- 50% of that course was theoretical, this will be reduced to 30% | ||
| − | Course website: <code>http://stab.st-andrews.ac.uk/i2rda/</code> | + | * Each section begins with a "Talk", and then a practical runthrough. |
| + | * If necessary, some talk slides may be skipped, as the main idea is getting through the practicals. | ||
| + | * Having said that if major theoretical points arise during a practical, they will be discussed. | ||
| + | * Course website: <code>http://stab.st-andrews.ac.uk/i2rda/</code> | ||
| + | :- this has all the presentations and practicals | ||
= Connecting to a remote Machine = | = Connecting to a remote Machine = | ||
| Line 18: | Line 23: | ||
==Configuring PuTTY for connection == | ==Configuring PuTTY for connection == | ||
| − | * Server: marvin.st-andrews.ac.uk | + | * Server: <code>marvin.st-andrews.ac.uk</code> |
* Terminal | keyboard | check VT100+ | * Terminal | keyboard | check VT100+ | ||
* Window | Selection | Control use of Mouse | set xterm | * Window | Selection | Control use of Mouse | set xterm | ||
| Line 26: | Line 31: | ||
You should now be able to "open" a session | You should now be able to "open" a session | ||
| − | * Be aware | + | * Be aware: typing in your password is done blindly. I.e. it does not appear on the screen. |
| − | + | Note: If you don't have your password, please ask to have it reset for you. | |
| − | + | = Computing resources notes = | |
| − | + | * RNA-Seq like other Next Generation Sequencing technologies, is characterised by; | |
| + | :- heavy computational workloads | ||
| + | :- many different software programs, sometimes doing the same thing, which can be arranged into a pipeline. | ||
| + | :- long-running tasks. | ||
| − | + | These have three implications: | |
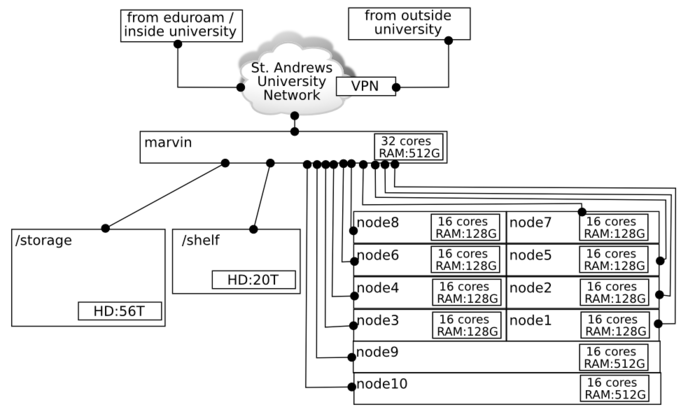
:- The marvin cluster is an 11-machine '''shared''' computing resource, not a personal computer ... others are using it. | :- The marvin cluster is an 11-machine '''shared''' computing resource, not a personal computer ... others are using it. | ||
| + | :- We need to load the special software before using it | ||
:- We want to be able to have a process run unattended. | :- We want to be able to have a process run unattended. | ||
| − | |||
| − | |||
| − | |||
| − | + | * For these three aspects, we have: | |
| + | :- A queue system to use, we shall request an interactive session ('''<code>qrsh</code>''') from the queue. | ||
| + | :- Use the ''module'' system to load, list and unload software programs | ||
| + | :- We shall use the '''GNU Screen''' utility so we can do other things while waiting. | ||
| − | + | = Computing resources diagram = | |
| − | To enter a new session | + | |
| − | + | [[File:marv.png]] | |
| − | * This will open with quite bare screen except with a bottom line | + | |
| − | * | + | = Aspects of using Windows terminals to connect to Linux = |
| − | * | + | |
| + | * You can pen the <code>http://st-andrews.ac.uk/i2rda</code> site on the Windows web-browser (Chrome preferred) and copy text selecting and <code>ctrl+c</code> | ||
| + | * This can then be pasted inside the PuTTY command-line by clicking the middle mouse button. | ||
| + | * In many ways, copy-pasting is not great for learning. | ||
| + | :- although some of the commands are too long to type out, even with history and tab-completion. | ||
| + | :- try to also use tab-completion, and the history (<code>up</code>/<code>down</code> arrows and <code>Ctrl+r</code>) | ||
| + | |||
| + | <ins>Weakness to watch out for</ins>: | ||
| + | * The marvin cluster (more precisely, the network it's attached to) doesn't carry graphics so well. | ||
| + | * We shall be using several graphical programs, and they are all likely to run slowly. | ||
| + | :- and sometimes even stall | ||
| + | :- we'll cross that bridge when we come to it. | ||
| + | |||
| + | =GNU Screen 1 = | ||
| + | |||
| + | A program which allows several command-line sessions open, similar to the idea of open tabs in a web browser. Let's try it out. | ||
| + | * To enter a new session, type <code>screen</code> | ||
| + | * This will open with quite a bare screen except with a indicator line at the bottom. | ||
| + | * <code>screen</code> works on the ''activator'' key concept, you need to use <code>Ctrl+l</code> (while <code>Ctrl</code>-key iis held down briefly, <code>l</code>-key is pressed) to activate any of its functions. | ||
| + | * After pressing <code>Ctrl+l</code> and releasing you then have a series of single key strokes that will do various useful things. | ||
| + | * There will be one command-line session open when you start it. | ||
| + | :- it's numbered 0, and called <code>scr1</code>, we'll mostly deal with it as <code>0</code>. | ||
| + | * Let's learn how to get out of it first | ||
| + | :- type <code>exit</code> again and you will be told you have exited <code>screen</code>. | ||
| + | :- you are now back in the ordinary command-line. | ||
| + | |||
| + | = GNU Screen 2 = | ||
| + | * Go back into screen, type <code>screen</code>. | ||
| + | * you open a new session with <code>ctrl+l,c</code> which '''c'''reates a new session. | ||
| + | :- you now have two sessions open | ||
| + | * type <code>ctrl+l,c</code> again, for three open sessions. You can have more, but we'll stick to three: number <code>0</code>, <code>1</code> and <code>2</code>. | ||
| + | * Switch back and forth between the three open sessions: use <code>Ctrl+l,n</code> ('''n''' for next) or <code>Ctrl+l,p</code> ('''p''' for previous) | ||
| + | * Don't see anything different when you do this? Look again at the bottom line, the asterisk has changed position. | ||
| + | :- the asterisk defines the active session | ||
| + | :- you can move to a numbered session directly with <code>ctrl+l,1</code> or <code>ctrl+0,1,2</code> for sessions 0, 1 and 2. | ||
| + | |||
| + | = Getting a Queue slot = | ||
| + | |||
| + | We're going to use one of the screen sessions to get a slot from the queue. | ||
| + | * Assuming you've launched screen, type <code>ctrl+l,0</code> to confirm you are in the first screen session. | ||
| + | * Type <code>qrsh</code> which requests a queue slot ... it will take a little time to give you one. | ||
| + | :- we shall not use this slot for the graphical programs, only the processing ones. | ||
| + | * When you get a slot, notice if you are still on marvin, or one of the nodes (assignment is based on load usually) | ||
| + | :- type qstat to see that you have allocated slot working in the queue. | ||
| + | * let's get something trivial running here: execute the <code>prtgn.sh</code> script by typing <code>prtgn.sh</code>, then <code>RETURN</code>, and let it print out gene names to its heart's content. | ||
| + | :- it's not drosophila, so only some of them are funny. | ||
| + | |||
| + | = Recovering a session = | ||
| + | |||
| + | * Now detach a session, <code>ctrl+l,d</code> to detach | ||
| + | * Now you're outside screen, you can log out, switch off and go home if you like (don't please). | ||
| + | * Next type <code>screen -r</code> to re-attach. | ||
| + | :- did the process stop? | ||
| + | :- unfortunately this cannot be done with many graphical programs | ||
| + | :- though some have a command-line mode, where it is possible | ||
| + | * Type ctrl+c, to stop it, demonstration over. | ||
| + | * you can also record the whole session, inputs and outputs in a file | ||
| + | :- done via <code>ctrl+l,:</code>, then type <code>hardcopy</code> and <code>RETURN</code> | ||
| + | :- the name of the file is <code>hardcopy.0</code> | ||
Latest revision as of 16:21, 11 May 2017
Contents
Course schedule
- This is based on a 2 day Edinburgh Genomics course of the same name, with the following changes:
- - "Introduction to Linux" moduel excluded
- - "Sequencer technology overview" module excluded.
- - No laboratory visit
- - 50% of that course was theoretical, this will be reduced to 30%
- Each section begins with a "Talk", and then a practical runthrough.
- If necessary, some talk slides may be skipped, as the main idea is getting through the practicals.
- Having said that if major theoretical points arise during a practical, they will be discussed.
- Course website:
http://stab.st-andrews.ac.uk/i2rda/
- - this has all the presentations and practicals
Connecting to a remote Machine
Presenting this before introduction as some people might experience delays logging in.
- We shall use a remote machine not the machine you are logged into locally
- The program we shall use is PuTTY.
- Please try to locate PuTTY in the applications section or on AppsAnywhere
Configuring PuTTY for connection
- Server:
marvin.st-andrews.ac.uk - Terminal | keyboard | check VT100+
- Window | Selection | Control use of Mouse | set xterm
- Connect | Data | enter username
- Connection | ssh | X11 Forwarding | Check yes
- Back to PuTTY main screen | select Default setting |click save
You should now be able to "open" a session
- Be aware: typing in your password is done blindly. I.e. it does not appear on the screen.
Note: If you don't have your password, please ask to have it reset for you.
Computing resources notes
- RNA-Seq like other Next Generation Sequencing technologies, is characterised by;
- - heavy computational workloads
- - many different software programs, sometimes doing the same thing, which can be arranged into a pipeline.
- - long-running tasks.
These have three implications:
- - The marvin cluster is an 11-machine shared computing resource, not a personal computer ... others are using it.
- - We need to load the special software before using it
- - We want to be able to have a process run unattended.
- For these three aspects, we have:
- - A queue system to use, we shall request an interactive session (
qrsh) from the queue. - - Use the module system to load, list and unload software programs
- - We shall use the GNU Screen utility so we can do other things while waiting.
Computing resources diagram
Aspects of using Windows terminals to connect to Linux
- You can pen the
http://st-andrews.ac.uk/i2rdasite on the Windows web-browser (Chrome preferred) and copy text selecting andctrl+c - This can then be pasted inside the PuTTY command-line by clicking the middle mouse button.
- In many ways, copy-pasting is not great for learning.
- - although some of the commands are too long to type out, even with history and tab-completion.
- - try to also use tab-completion, and the history (
up/downarrows andCtrl+r)
Weakness to watch out for:
- The marvin cluster (more precisely, the network it's attached to) doesn't carry graphics so well.
- We shall be using several graphical programs, and they are all likely to run slowly.
- - and sometimes even stall
- - we'll cross that bridge when we come to it.
GNU Screen 1
A program which allows several command-line sessions open, similar to the idea of open tabs in a web browser. Let's try it out.
- To enter a new session, type
screen - This will open with quite a bare screen except with a indicator line at the bottom.
-
screenworks on the activator key concept, you need to useCtrl+l(whileCtrl-key iis held down briefly,l-key is pressed) to activate any of its functions. - After pressing
Ctrl+land releasing you then have a series of single key strokes that will do various useful things. - There will be one command-line session open when you start it.
- - it's numbered 0, and called
scr1, we'll mostly deal with it as0.
- Let's learn how to get out of it first
- - type
exitagain and you will be told you have exitedscreen. - - you are now back in the ordinary command-line.
GNU Screen 2
- Go back into screen, type
screen. - you open a new session with
ctrl+l,cwhich creates a new session.
- - you now have two sessions open
- type
ctrl+l,cagain, for three open sessions. You can have more, but we'll stick to three: number0,1and2. - Switch back and forth between the three open sessions: use
Ctrl+l,n(n for next) orCtrl+l,p(p for previous) - Don't see anything different when you do this? Look again at the bottom line, the asterisk has changed position.
- - the asterisk defines the active session
- - you can move to a numbered session directly with
ctrl+l,1orctrl+0,1,2for sessions 0, 1 and 2.
Getting a Queue slot
We're going to use one of the screen sessions to get a slot from the queue.
- Assuming you've launched screen, type
ctrl+l,0to confirm you are in the first screen session. - Type
qrshwhich requests a queue slot ... it will take a little time to give you one.
- - we shall not use this slot for the graphical programs, only the processing ones.
- When you get a slot, notice if you are still on marvin, or one of the nodes (assignment is based on load usually)
- - type qstat to see that you have allocated slot working in the queue.
- let's get something trivial running here: execute the
prtgn.shscript by typingprtgn.sh, thenRETURN, and let it print out gene names to its heart's content.
- - it's not drosophila, so only some of them are funny.
Recovering a session
- Now detach a session,
ctrl+l,dto detach - Now you're outside screen, you can log out, switch off and go home if you like (don't please).
- Next type
screen -rto re-attach.
- - did the process stop?
- - unfortunately this cannot be done with many graphical programs
- - though some have a command-line mode, where it is possible
- Type ctrl+c, to stop it, demonstration over.
- you can also record the whole session, inputs and outputs in a file
- - done via
ctrl+l,:, then typehardcopyandRETURN - - the name of the file is
hardcopy.0