Difference between revisions of "Velvet"
(Created page with "This is Daniel Zerbino and Ewan Birney's de-novo genome assembler and is one of the most widely used. It consists of two stages, a hashing stage invoked by the velveth progra...") |
|||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
This is Daniel Zerbino and Ewan Birney's de-novo genome assembler and is one of the most widely used. | This is Daniel Zerbino and Ewan Birney's de-novo genome assembler and is one of the most widely used. | ||
| − | It consists of two stages, a hashing stage invoked by the velveth program, and then the actual genome assembly stage | + | It consists of two stages, a hashing stage invoked by the '''velveth''' program, and then the actual genome assembly stage |
| − | which is invoked by the velvetg program. | + | which is invoked by the '''velvetg''' program. |
| − | As with many other genome assemblers, the program splits short reads into shorter sequences of | + | As with many other genome assemblers, the program splits short reads into even shorter sequences of length k, called kmers and |
arranges these in de Bruijn graphs from which the genome is then assembled. | arranges these in de Bruijn graphs from which the genome is then assembled. | ||
| + | |||
| + | == input fastq datasets == | ||
| + | |||
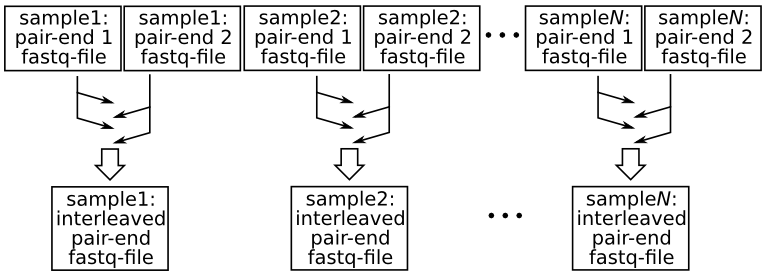
| + | Velvet differs from other assemblers in requiring that the input short reads, if consisting of pair-end reads, be provided within a single file rather than the more usual two files (i.e. one file for each member of the pair). For Velvet, the two member-reads of each corresponding pair need to follow each other within a single file in what may be called an interleaved, or shuffled, format. The following diagram illustrates the procedure: | ||
| + | |||
| + | [[File:intleaved.png]] | ||
| + | |||
| + | = Installation = | ||
| + | |||
| + | Velvet has a few options that are useful to build in at compile time. For colorspace aeembly you will need special executables, ending in "de" and they are compiled with | ||
| + | make color | ||
| + | |||
| + | For allowing a greater variation of kmer sizes (default is a rather lowly 31), ht efollowing compile-time option is avilable. | ||
Latest revision as of 17:14, 19 December 2016
This is Daniel Zerbino and Ewan Birney's de-novo genome assembler and is one of the most widely used.
It consists of two stages, a hashing stage invoked by the velveth program, and then the actual genome assembly stage which is invoked by the velvetg program.
As with many other genome assemblers, the program splits short reads into even shorter sequences of length k, called kmers and arranges these in de Bruijn graphs from which the genome is then assembled.
input fastq datasets
Velvet differs from other assemblers in requiring that the input short reads, if consisting of pair-end reads, be provided within a single file rather than the more usual two files (i.e. one file for each member of the pair). For Velvet, the two member-reads of each corresponding pair need to follow each other within a single file in what may be called an interleaved, or shuffled, format. The following diagram illustrates the procedure:
Installation
Velvet has a few options that are useful to build in at compile time. For colorspace aeembly you will need special executables, ending in "de" and they are compiled with
make color
For allowing a greater variation of kmer sizes (default is a rather lowly 31), ht efollowing compile-time option is avilable.