Difference between revisions of "RNASeq library preparation"
(Created page with "= Total RNA = File:totalrna.png = mRNA-seq workflow = File:rnawf.png = Illumina NGS Library = File:illu.png = Illumina library read binding 1= File:illu2....") |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 11: | Line 11: | ||
[[File:illu.png]] | [[File:illu.png]] | ||
| − | = Illumina library | + | = Illumina library fragment = |
[[File:illu2.png]] | [[File:illu2.png]] | ||
| − | = Illumina library read | + | = Illumina library read bindings 1= |
[[File:illu4.png]] | [[File:illu4.png]] | ||
| − | = Illumina library read | + | = Illumina library read bindings 2= |
[[File:illu5.png]] | [[File:illu5.png]] | ||
| Line 42: | Line 42: | ||
[[file:dan.png]] | [[file:dan.png]] | ||
| + | |||
| + | = The number of replicates issue = | ||
| + | [[File:rnasoc.png]] | ||
| + | * http://rnajournal.cshlp.org/content/22/6/839.short | ||
| + | [[FIle:biorpaper.png]] | ||
| + | * http://www.biorxiv.org/content/early/2016/12/02/090753 | ||
Latest revision as of 10:27, 9 May 2017
Contents
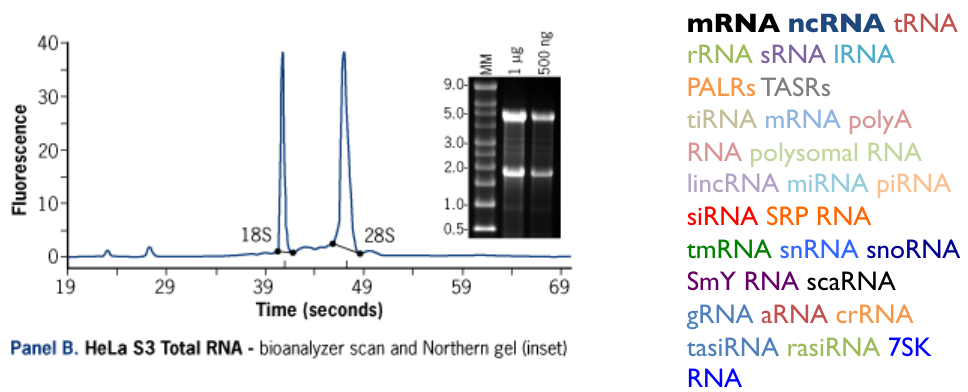
Total RNA
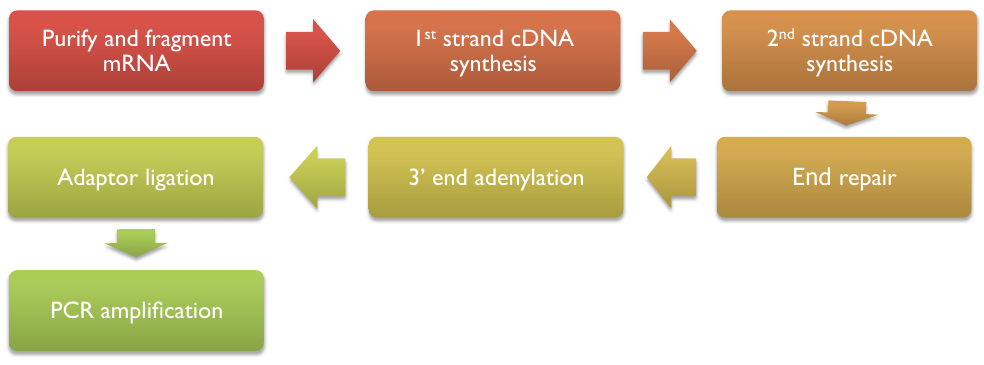
mRNA-seq workflow
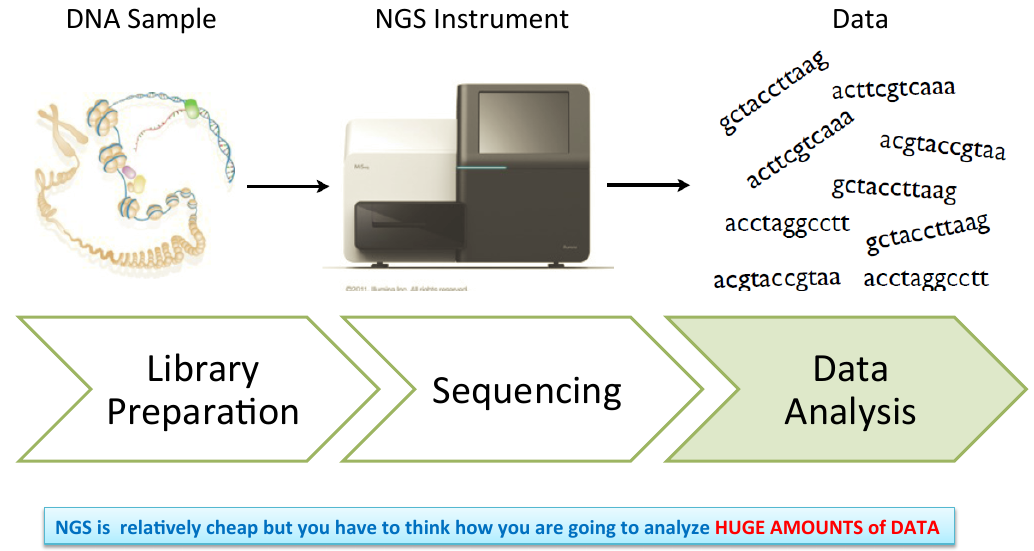
Illumina NGS Library
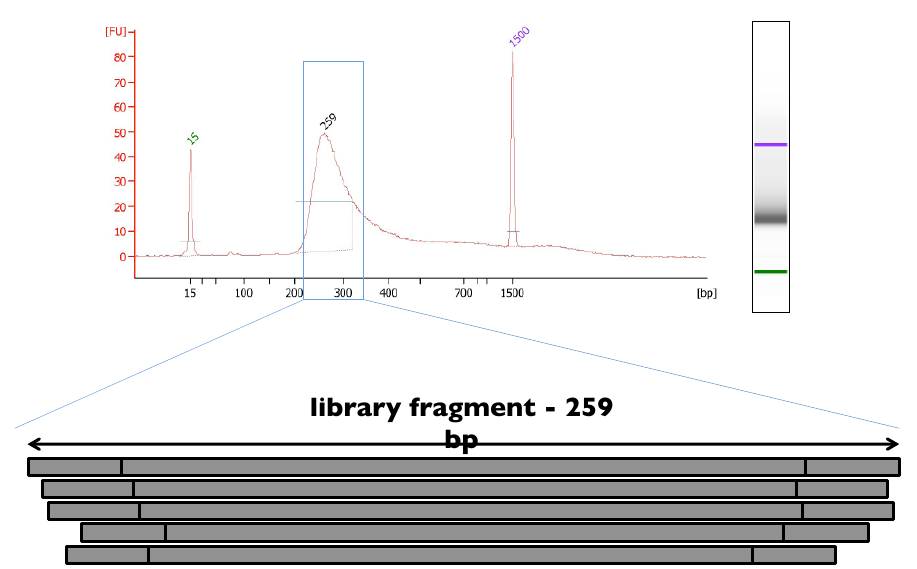
Illumina library fragment
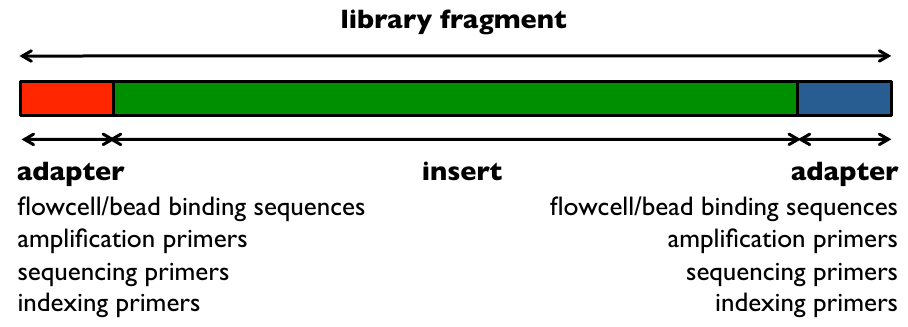
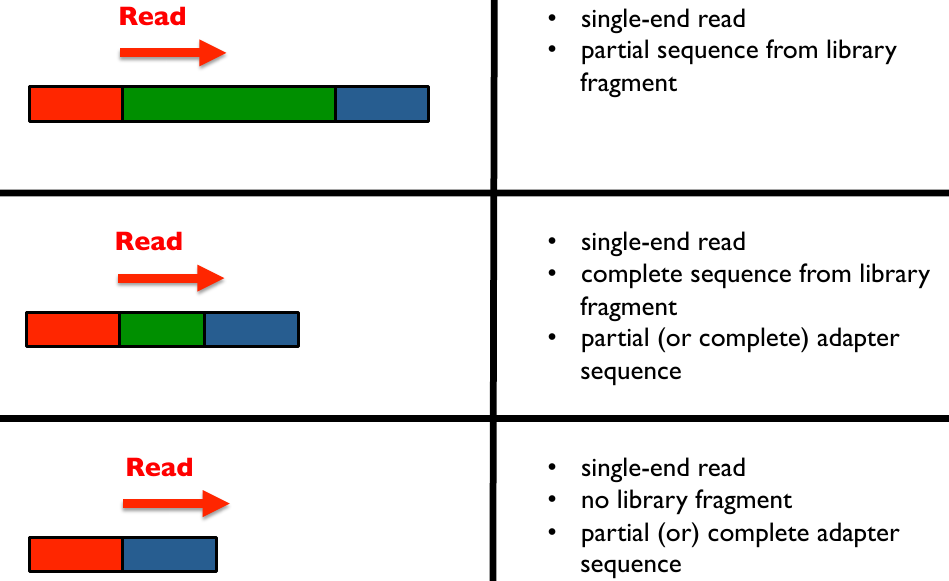
Illumina library read bindings 1
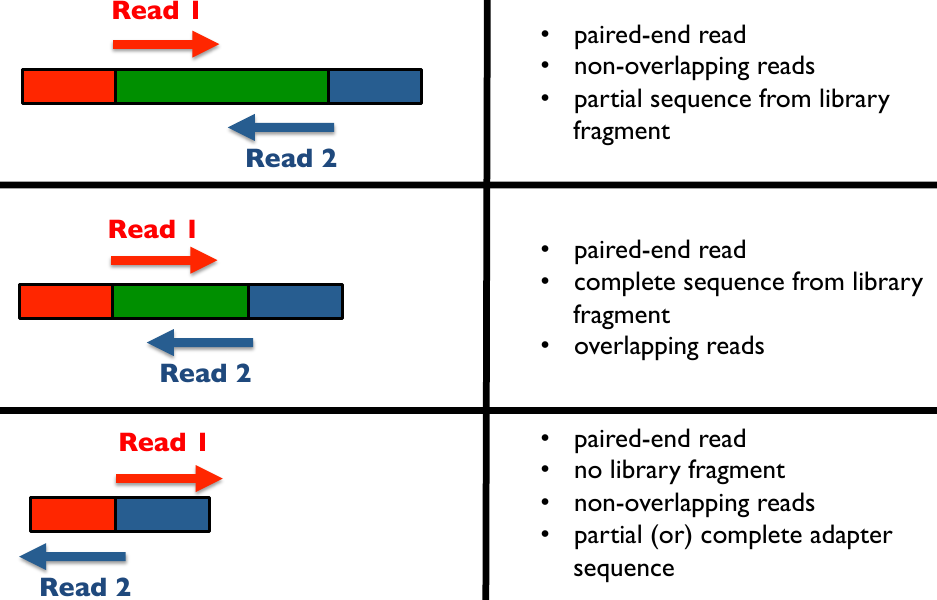
Illumina library read bindings 2
RNA-Seq applications
- rRNA depleted samples
- Very high dynamic range
- No prior knowledge of expressed genes.
- - Gives more information than microarrays.
- Applications
- - Sequencing of mRNA
- - Differential expression of known or unknown transcripts during a treatment or condition
- - Isoform detection.
- - Alternative splicing events
- - Post-transcriptional mutations or editing
- - Gene fusions
- - Non-coding RNAs
- - miRNAs/small non-Coding RNA