Difference between revisions of "Synthetic Long reads"
| (One intermediate revision by the same user not shown) | |||
| Line 7: | Line 7: | ||
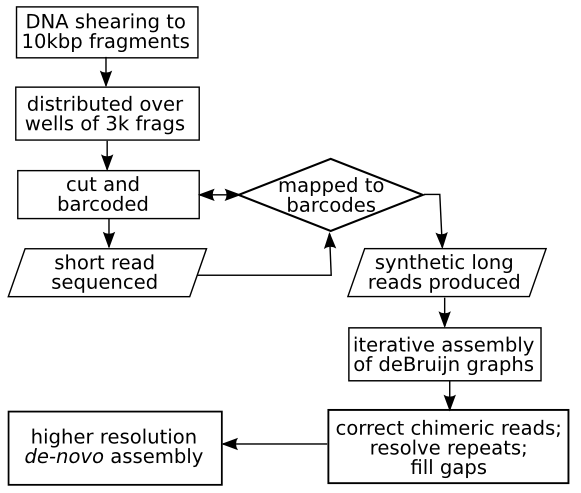
It is already installed on the Bioinformatics cluster to ensure fast processing. The pipeline starts by synthesising long reads by mapping the short read to the barcoded pools. Next, a de Bruijn graph - a compact representation of the long reads in their order of sequence - is iteratively refined into scaffolds through a series of error-correction and coverage gap-filling stages. The resulting de-novo assembly is of higher resolution than can be achieved with a higher percentage of spanned gaps and errors corrected. | It is already installed on the Bioinformatics cluster to ensure fast processing. The pipeline starts by synthesising long reads by mapping the short read to the barcoded pools. Next, a de Bruijn graph - a compact representation of the long reads in their order of sequence - is iteratively refined into scaffolds through a series of error-correction and coverage gap-filling stages. The resulting de-novo assembly is of higher resolution than can be achieved with a higher percentage of spanned gaps and errors corrected. | ||
| − | + | The general procedure for synthetic long reads is as follows: | |
| − | + | [[File:biopipe.png]] | |
Latest revision as of 16:02, 8 April 2016
NGS reads are typically short in that they found in the 50-200 base-pair length scale. Longer reads are seen as more powerful, so there is a general tendency towards longer reads which is reflected in prices. Illumina generally have focussed on shorter reads, but in an effort to follow trends, they have the developed the synthetic long read.
A recent development, synthetic long reads have already been used successfully in projects for complex transposon resolution, recovery of missing sequences, metagenomics and exome enrichment.
The goal of the bioinformatics pipeline is to produce a de-novo assembly of higher resolution where sequencing gaps can be correctly bridged by virtue of the increase read length. One software that can be used is the enhancement of the widely used SPAdes program called TruSPAdes[1] which is focused on handling the particular challenge of assembling synthetic long reads, such as handling and correcting chimeric reads
It is already installed on the Bioinformatics cluster to ensure fast processing. The pipeline starts by synthesising long reads by mapping the short read to the barcoded pools. Next, a de Bruijn graph - a compact representation of the long reads in their order of sequence - is iteratively refined into scaffolds through a series of error-correction and coverage gap-filling stages. The resulting de-novo assembly is of higher resolution than can be achieved with a higher percentage of spanned gaps and errors corrected.
The general procedure for synthetic long reads is as follows:
- ↑ Bankevich, A. and Pevzner, P. A. (2016) TruSPAdes: barcode assembly of TruSeq synthetic long reads, Nature Methods 3: 248-250