Difference between revisions of "MinION (Oxford Nanopore)"
| Line 16: | Line 16: | ||
* Reads an be quite long ... 100kb is possible. | * Reads an be quite long ... 100kb is possible. | ||
| − | [[pore0.png]] | + | [[File:pore0.png]] |
==Shortcomings== | ==Shortcomings== | ||
| Line 28: | Line 28: | ||
* DNA prep about 2 hours, but a "rapid kit" exists which makes 10 minutes possible. | * DNA prep about 2 hours, but a "rapid kit" exists which makes 10 minutes possible. | ||
| − | [[prep0.png]] | + | [[File:prep0.png]] |
= Software Round-up = | = Software Round-up = | ||
Revision as of 13:47, 2 February 2017
Contents
Introduction
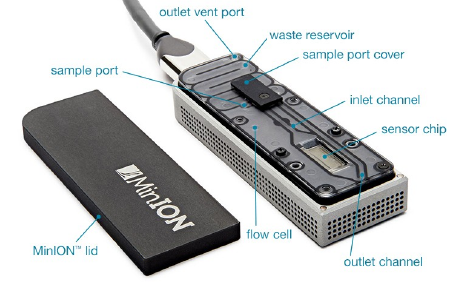
Entirely unique sequencing method, where the flowcell is inserted into a USB container, and from there, plugged into a computer.
Remarkable due to its size when it was first announced aroudn 2010, it grabbed all the headlines. However, its creator, Oxford Nanopore Technologies (ONT for short) has consistently overpromised on its capabilities, though this is entirely normal for a commercial company which seeks investors early.
Due to its small size in comparison with Illumina, IonTorrent and PacBio, this sequencing tool is eminently suited to field work.
Overview
Reputed advantages
- flowcell pores good for several runs, until they die out, which they may do at different times.
- Reads an be quite long ... 100kb is possible.
Shortcomings
- Computer, usually a laptop, needs to be continually connected to internet, and to be in high workload mode (no economy nor sleep mode allowed).
- accuracy at least an order of magnitude worse than Illumina (~90% vs >99%)
- Probably more expensive than Illumina on a per-base basis, although there is no service contract involved as one might expect from Illumina. Low cost of Illumina cost is largely down to economies of scale.
Characteristics
- 1D, which means single-strand reading, is the most common and mature of MinION's modes.
- 2D, where both strands are read, one after the other, is possible, and allows much better accuracy, but is more demanding and more prone to errors.
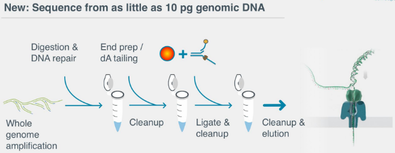
- DNA prep about 2 hours, but a "rapid kit" exists which makes 10 minutes possible.
Software Round-up
The software required can be split into two groups of programs:
- Sequencing generation
- MinKNOW, for control of MinION device & run parameters
- Metrichor, for cloud basecalling of event data
- Chronolapse a screen image grabber for record keeping
- TeamViewer, for remote control of MinION computer
- MinoTour, live monitoring / control of run while sequencing (a collaboration with Matt Loose of Nottingham University).
The main product of these tools is the fast5 file format. This format has good metadata capabilities, though the extent and usefulness of this metadata depends on how the experiment is run.
- Sequence File Analysis
- Poretools, poRe Sequence extraction and data summaries (deevloped by Nick Loman and Aaron Quinlan (latter of bedtools fame)).
Examples of poretools usage
poretools fastq 5CG6210Y8Z_20160816_FNFAB28012_MN15120_sequencing_run_GroupB_1D_Ecoli_tune_85746_ch39_read230_strand.fast5
Explanation:
- poretools is the main tool command
- fastq is the subcommand, and it mostly defines the output that the user requires. The input is expected to be a list of fast5 filenames or a directory
- the rest of the command is actually just the fast5 filename, which is clearly very long, and is probably due to the high metadata capabilities of fast5
Developments during 2016
At the beginning of 2016, udring a regular run, MinION could:
- Process 500Mb of DNA from a flow-cell
- Each pore could read 70 bases/second
- Accuracy still low at 70-80%
Major steps were made to improve this:
- Pore technology moved from R7 (had patent conflict problems with Illumina) to R9 (better throughput, higher accuracy). R9 itself is continuously being incrementally improved. As of Jan 2017 it was at version R9.4.
- implemented new deep-learning algorithm
- closer collaboration with key academic researchers.
Links
- Brian Naughton's blog entry 11 Oct 2016 taking stock of recent advances
- Nature paper 11 Feb 2016 describing Minion use in Ebola outbreak