Difference between revisions of "MinION (Oxford Nanopore)"
| Line 23: | Line 23: | ||
* '''2D''', where both strands are read, one after the other, is possible, and allows much better accuracy, but is more demanding and more prone to errors. | * '''2D''', where both strands are read, one after the other, is possible, and allows much better accuracy, but is more demanding and more prone to errors. | ||
| − | + | = Software Round-up = | |
The software required can be split into two groups of programs: | The software required can be split into two groups of programs: | ||
Revision as of 11:28, 2 February 2017
Contents
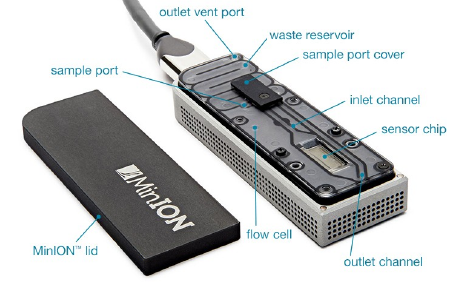
Introduction
Entirely unique sequencing method, where the flowcell is inserted into a USB container, and from there, plugged into a computer.
Due to its small size in comparison with Illumina, IonTorrent and PacBio, this sequencing tool is eminently suited to field work.
Overview
Reputed advantages
- flowcell pores good for several runs, until they die out, which they may do at different times.
- Reads an be quite long ... 100kb is possible.
Shortcomings
- Computer, usually a laptop, needs to be continually connected to internet, and to be in high workload mode (no economy nor sleep mode allowed).
- accuracy at least an order of magnitude worse than Illumina (~90% vs >99%)
- Probably more expensive than Illumina on a per-base basis, although there is no service contract involved as one might expect from Illumina. Low cost of Illumina cost is largely down to economies of scale.
Characteristics
- 1D, which means single-strand reading, is the most common and mature of MinION's modes.
- 2D, where both strands are read, one after the other, is possible, and allows much better accuracy, but is more demanding and more prone to errors.
Software Round-up
The software required can be split into two groups of programs:
- Sequencing generation
- MinKNOW, for control of MinION device & run parameters
- Metrichor, for cloud basecalling of event data
- Chronolapse a screen image grabber for record keeping
- TeamViewer, for remote control of MinION computer
- MinoTour, live monitoring / control of run while sequencing (a collaboration with Matt Loose of Nottingham University).
- Sequence File Analysis
- Poretools, poRe Sequence extraction and data summaries (deevloped by Nick Loman and Aaron Quinlan (latter of bedtools fame)).
Developments during 2016
At the beginning of 2016, udring a regular run, MinION could:
- Process 500Mb of DNA from a flow-cell
- Each pore could read 70 bases/second
- Accuracy still low at 70-80%
Major steps were made to improve this:
- Pore technology moved from R7 (had patent conflict problems with Illumina) to R9 (better throughput, higher accuracy)
- implemented new deep-learning algorithm
- closer collaboration with key academic researchers.
Links
- Brian Naugton's blog entry 11 Oct 2016 taking stock of recent advances
- Nature paper 11 Feb 2016 describing Minion use in Ebola outbreak