Difference between revisions of "MinION (Oxford Nanopore)"
| (2 intermediate revisions by the same user not shown) | |||
| Line 77: | Line 77: | ||
* It seems that MinKNOW needs the Metrichor agent to be running, although this is not entirely clear. | * It seems that MinKNOW needs the Metrichor agent to be running, although this is not entirely clear. | ||
* Traditionally they require Windows. But it both packages are now avilable for MacOSX. | * Traditionally they require Windows. But it both packages are now avilable for MacOSX. | ||
| − | * ONT also have the MinKNOW software for Linux, namely Ubuntu Trusty 14.04 (which actually is the latest Bio Linux installation) but there is no Ubuntu version of Metrichor, so this does not seem useful. | + | * ONT also have the MinKNOW software for Linux, namely Ubuntu Trusty 14.04 (which actually is the latest Bio Linux installation) but there is no Ubuntu version of Metrichor, so this does not seem useful. However on Wed 8 Feb 2017, and Ubuntu version of the user agent for Ubuntu was released. |
* Metrichor has its own website at www.metrichor.com. Authentication seems to be done via the nanopore website however. | * Metrichor has its own website at www.metrichor.com. Authentication seems to be done via the nanopore website however. | ||
| Line 87: | Line 87: | ||
* '''Poretools''', poRe Sequence extraction and data summaries (developed by Nick Loman and Aaron Quinlan (latter of bedtools fame)). This is installed on the marvin cluster with version number 0.6.0 as of Jan 2017. | * '''Poretools''', poRe Sequence extraction and data summaries (developed by Nick Loman and Aaron Quinlan (latter of bedtools fame)). This is installed on the marvin cluster with version number 0.6.0 as of Jan 2017. | ||
* '''poRe''', by Mick Watson, very similar to poretools, but for the R statistics platform. Found [https://sourceforge.net/projects/rpore/files/0.20 here] | * '''poRe''', by Mick Watson, very similar to poretools, but for the R statistics platform. Found [https://sourceforge.net/projects/rpore/files/0.20 here] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
= Links = | = Links = | ||
| Line 129: | Line 98: | ||
* [https://github.com/mw55309/EG_MinION_2016/blob/master/02_Data_Extraction_QC.md key aspects] of the fast5 file format | * [https://github.com/mw55309/EG_MinION_2016/blob/master/02_Data_Extraction_QC.md key aspects] of the fast5 file format | ||
* [http://porecamp.github.io/2016/ Porecamp 2016, training course page] | * [http://porecamp.github.io/2016/ Porecamp 2016, training course page] | ||
| + | * [https://community.nanoporetech.com/posts/command-line-agent-release October 2017 latest software link] | ||
=Notes= | =Notes= | ||
<references /> | <references /> | ||
Latest revision as of 09:55, 17 October 2017
Contents
Introduction
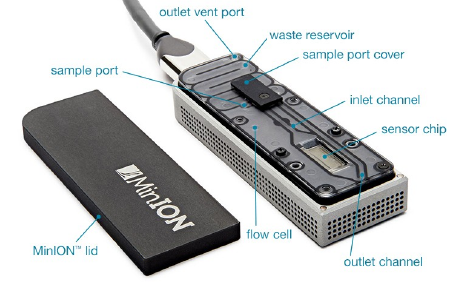
Entirely unique sequencing method, where the flowcell is inserted into a USB container, and from there, plugged into a computer. Its creator, Oxford Nanopore Technologies (ONT for short) is a commercial company.
Remarkable due to its size when it was first announced at the "Advances in Genome Biology and Technology" in 2012 by Clive G. Brown (CEO of ONT) and it grabbed all the headlines. However, ONT has consistently overpromised on its capabilities (the MinION was oly aviilable two years later), though this is entirely normal for a commercial company which seeks investors early. Though early improvements were slow, sometimes not providing enough data for genome assembly [1], during 2016 the technology seemed to be reaching a certain maturity both due to key improvements and successful trials of the technology presented by the Ebola outbreak and the Zika virus projects.
Due to its small size in comparison with Illumina, IonTorrent and PacBio, this sequencing tool is eminently suited to field work.
Overview
Reputed advantages
- flowcell pores good for several runs, until they die out, which can be in the order of 48 hours.
- Reads an be quite long ... 100kb is possible.
Shortcomings
- Computer, usually a laptop, needs to be continually connected to internet, and to be in high workload mode (no economy nor sleep mode allowed).
- accuracy at least an order of magnitude worse than Illumina (~90% vs >99%)
- Probably more expensive than Illumina on a per-base basis, although there is no service contract involved as one might expect from Illumina. Low cost of Illumina cost is largely down to economies of scale.
Characteristics
- 1D, which means single-strand reading, is the most common and mature of MinION's modes.
- 2D, where both strands are read, one after the other, is possible, and allows much better accuracy, but is more demanding and more prone to errors.
- DNA prep about 2 hours, but a "rapid kit" exists which makes 10 minutes possible. For 1D 15 minutes is also doable.
Broad Explanation of pore base-calling method
- DNA passes through the pore: changes in ionic current detected
- These changes caused by differences in the shifting nucleotide sequences occupying the pore.
- Changes segmented as discrete events that have an associated duration, mean amplitude, and variance.
- Sequence of events interpreted computationally as a sequence of 3–6 nucleotide long kmers (‘words’) using graphical models.
- Information from template and complement reads is combined to produce a high-quality ‘2D read’, using a pairwise alignment of the event sequences.
More detail can be found here [2]
Developments during 2016
At the beginning of 2016, udring a regular run, MinION could:
- Process 500Mb of DNA from a flow-cell
- Each pore could read 70 bases/second
- Accuracy still low at 70-80%
Major steps were made to improve this:
- Pore technology moved from R7 (had patent conflict problems with Illumina) to R9 (better throughput, higher accuracy).
- R9 itself is continuously being incrementally improved. First version: 270bps, 85% 1D identity (95% 2D identityt)
- As of Jan 2017 it was at version R9.4 capable of 450bps at 1D with 90% identity.
- New large device developed: PromethION with 48 flowcells.
- implemented new deep-learning algorithm
- closer collaboration with key academic researchers.
Use Cases
- An example with detailed steps of a MinION run can be found here.
- Josh Quick's presentation about in-the-field usage of MinION
- Gene mutation analysis, an example of targeted use of MinION [3]
Software Round-up
The software required can be split into two groups of programs:
Sequencing generation
- MinKNOW, for control of MinION device & run parameters
- Metrichor, for cloud basecalling of event data
- Chronolapse a screen image grabber for record keeping
- TeamViewer, for remote control of MinION computer
- MinoTour, live monitoring / control of run while sequencing (a collaboration with Matt Loose of Nottingham University).
- MinUP, a Matt Loose tool allowing uploading of data for the MinoTour program
MinKNOW and Metrichor
- These are the core ONT programs.
- It seems that MinKNOW needs the Metrichor agent to be running, although this is not entirely clear.
- Traditionally they require Windows. But it both packages are now avilable for MacOSX.
- ONT also have the MinKNOW software for Linux, namely Ubuntu Trusty 14.04 (which actually is the latest Bio Linux installation) but there is no Ubuntu version of Metrichor, so this does not seem useful. However on Wed 8 Feb 2017, and Ubuntu version of the user agent for Ubuntu was released.
- Metrichor has its own website at www.metrichor.com. Authentication seems to be done via the nanopore website however.
The main product of these tools is the fast5 file format. This format has good metadata capabilities, though the extent and usefulness of this metadata depends on how the experiment is run.
However, direct monitoring of the base calls is also now possible, due to Matt Loose at Nottingham University, using the MinoTour platform. This requires an external server, one of which is available at University of Nottingham, but it is also possible to set one up internally if need be.
Sequence File Analysis
- Poretools, poRe Sequence extraction and data summaries (developed by Nick Loman and Aaron Quinlan (latter of bedtools fame)). This is installed on the marvin cluster with version number 0.6.0 as of Jan 2017.
- poRe, by Mick Watson, very similar to poretools, but for the R statistics platform. Found here
Links
General Minion Procedures
Analysis Software Links
- Brian Naughton's blog entry 11 Oct 2016 taking stock of recent advances
- Nature paper 11 Feb 2016 describing Minion use in Ebola outbreak
- key aspects of the fast5 file format
- Porecamp 2016, training course page
- October 2017 latest software link
Notes
- ↑ Judith Risse, Marian Thomson, Sheila Patrick, Garry Blakely, Georgios Koutsovoulos, Mark Blaxter and Mick Watson A single chromosome assembly of Bacteroides fragilis strain BE1 from Illumina and MinION nanopore sequencing data GigaScience 2015 4:60
- ↑ Miten Jain Hugh E. Olsen, Benedict Paten and Mark Akeson The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics communityGenome Biology 2016 17:239
- ↑ Crescenzio Francesco Minervini, Cosimo Cumbo, Paola Orsini, Claudia Brunetti, Luisa Anelli, Antonella Zagaria, Angela Minervini, Paola Casieri, Nicoletta Coccaro, Giuseppina Tota, Luciana Impera, Annamaria Giordano, Giorgina Specchia and Francesco Albano TP53 gene mutation analysis in chronic lymphocytic leukemia by nanopore MinION sequencing Diagnostic Pathology 2016 11:96