Available Software
Contents
Introduction
We now highly recommend conda to install your packages:
Conda. THIS IS THE FUTURE. We highly recommend this
We are testing Bioconda on Marvin on a per user basis, so you have total control over it.
1) Log in and from the command line type (only do this once!):
install-bioconda
2) Either log out and back in or type:
source ~/.bashrc
3) We strongly advise using environments: https://conda.io/docs/user-guide/tasks/manage-environments.html . You can do this in many ways. Please see the link for more details (here you can specifiy eact version etc ..). The easiest usage would be: e.g. conda create -n NAME_OF_ENV PACKAGE_TO_INSTALL (pick a package you are interested in from here: installing packages: https://bioconda.github.io/recipes.html ). Roary is a package name as an example.
conda create -n roary roary
conda activate roary
conda update roary
You are now ready to use this package.
Conda deactivate to leave this environment.
This works for me and a test user. But may fail for you, so be patient with us.
To list all the environments you have created:
conda info –envs
installing packages:
https://bioconda.github.io/recipes.html
module load approach
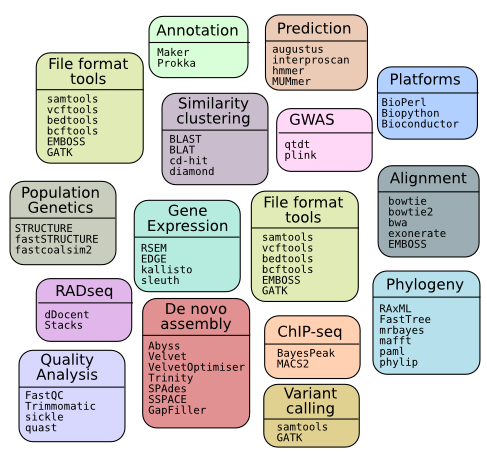
The cluster uses the widely used Environment Modules system to manage the wide range of software available for Bioinformatics. It attempts to take care of the necessary environment for running the software, and so minmise (although not erradicate) errors.
To see all the available software, the command to type is:
moduleav
This is a fast wrapper command for the underlying slower:
module available
Categorised Listing
Raw Listing 07 Dec 2016
--------------------------------- /usr/local/Modules/versions ----------------------------------------------------- 3.2.10 ---------------------------- /usr/local/Modules/3.2.10/modulefiles ------------------------------------------------ AdapterRemoval/gitv0_abe4a3e(default) ArtificialFastqGenerator/1.0.0(default) AsperaConnect/3.6.2 BAPS/6.0 BUSCO/1.22 BamQC/gitv0_480c091 BayeScEnv/1.0 BayeScEnv/gitv0_beb60d2(default) BayeScan/2.1(default) CAFE/3.1 CEGMA/2.5(default) CRISP/2 CRISP/5 CRISP/gitv0_7bfcd34(default) EIGENSOFT/6.0.1(default) EMBOSS/6.6.0(default) FASTQC/0.11.5(default) FastML/3.1 FastML/gitv0_bb5d761 FastTree/2.1.8(default) GapFiller/v1-10 GeneMark_ES/3.48(default) HTSlib/1.3 HTSlib/1.3.1(default) HTSlib/gitv1_0de7fe5 MCR/v713 MUMmer/3.23 MUMmer/3.23b(default) Mash/gitv0_2edc42 Mash/v1.1.1(default) Mauve/2015-02-13 NGSToolsApp/2.1.5(default) OpenBLAS/0.2.14 OpenBLAS/0.2.18 OpenBLAS/0.2.19(default) OrthoFinder/0.6.0 PAGIT/1(default) PEAR/gitv0_a37a84b(default) PEAT/gitv0_27981d7(default) PGAP/1.2.1 PGDSpider/2.0.8.2(default) Prodigal/gitv0_004218f R/3.2.1(default) R/3.2.3 R/3.3.1 RAxML/gitv0_4692545(default) RSEM/1.2.25(default) RSEM/1.2.27 SPAdes/3.9.0(default) SSPACE/3.0 STAR/2.4.2a(default) T-COFFEE/11.00.8 VarScan/2.3.7(default) VelvetOptimiser/2.2.5(default) abacas/1.3.1(default) abyss/1.9.0 act/v1 allpathslg/52488 angsd/gitv0_212d58 angsd/gitv1_89d8fa9(default) arlequin/3.5.2(default) artemis/16.0.0(default) assembly-stats/gitv0_003a372 augustus/3.0.3 augustus/3.2.2(default) bamtools/gitv0_02c2be8(default) bcftools/gitv0_47e811c bedtools/2.23.0 bedtools/2.25.0(default) bioawk/gitv0_5e8b41d(default) blastScripts/1.0.0(default) blastScripts/1.0.1 blastall/2.2.26(default) blat/35 boost/1.60.0(default) boost/1.61.0 bowtie/1.0.0 bowtie2/2.1.0(default) bowtie2/2.2.4 bwa/0.7.12(default) bwa/0.7.7 bzip2/1.0.6 capnproto/0.5.3 cd-hit/gitv0_5acf038(default) clustalx/2.1 crimap/2.504(default) cufflinks/2.2.1(default) dDocent/gitv0_7dea314 diamond/gitv0_39f2a71 dot ea-utils/gitv0_88c9ad5 ensembl-git-tools/gitv0_68ac669(default) ensembl/gitv0_bb48912(default) exonerate/2.2.0(default) fastSTRUCTURE/1.0 fastSTRUCTURE/190315(default) fastsimcoal2/2.5.2.21 fastsimcoal2/2.5.2.8(default) freebayes/gitv0_dbb6160 gatk/3.4.0 gatk/3.5.0 gatk/3.6.0(default) gcc/4.9.3 geneid/1.4.4(default) general_script_tools/(default) gnuparallel/20160422 gsea/2.2.2(default) gsl/2.1 gubbins/gitv0_fe094b0 gubbins/gitv1_8a12ae7(default) gwas harvest-tools/gitv0_ebf7ecc hdf5/1.8.15-patch1(default) hdf5/1.8.16 hmmer/2.3.2 hmmer/3.1b1 hmmer/3.1b2(default) interproscan/5.4-47.0(default) kallisto/git0_f0678a2 lamarc/2.1.10(default) lpsolve/5.5.2.3 mafft/7.147(default) maker/2.31.8(default) masurca/2.3.2(default) mcl/14-137(default) mdb/0.6(default) merlin/1.1.2(default) module-info modules mpifast/gitrev2(default) mrbayes/3.2.6 muscle/3.7 newick_utils/4.9.3 newick_utils/gitv0_614bf6c(default) ngs/gitv0_aef5f50 null openmpi/1.10.2 openmpi/1.6.5 openmpi/2.0.0(default) oraclejava/jdk1.8.0_74 paml/4.7a(default) parsnp/gitv0_63bea13 pblat/1.6.0(default) perl/5.20.2(default) perl/5.24.0 phylip/3.696(default) phyml/gitv0_0f6d745 picard-tools/1.119 picard-tools/2.1.0 picard-tools/2.1.1 picard-tools/2.7.1(default) prank-msa/gitv0_3e44771(default) prokka/1.11 prokka/gitv1_8f07048(default) prosite/1.79.1 protobuf/gitv0_0622030 psmc/gitv0_e5f7df5(default) pyrad/gitv0_704ffbf python/2.7(default) python/2.7.12 python/3.4 qtdt/2.6.1(default) quast/4.1 rainbow/2.0.4 rapsearch/gitv0_95c866e(default) readseq/2.1.30(default) repeatmasker/4.0.5(default) rmblastn/2.2.28(default) rnammer/1.2(default) roary/gitv0_c4d9b77(default) samtools/0.1.18 samtools/0.1.19(default) samtools/0.1.19b samtools/1.2 samtools/1.3.1 scalaBLAST/2.0(default) scipio/1.4.1(default) script_tools/1.0(default) seqtk/gitv0_1a8319b(default) sickle/gitv0_f3d6ae3(default) signalp/4.1(default) smalt/0.7.6(default) snap_alignment/1.0beta.17(default) snap_hmm/11_29_2013(default) snape-pooled/31(default) snippy/gitv0_848a62e snpcallphylo/0.7 sparsehash/gitv0_4cb9240 sra-tools/gitv0_814d6f7(default) srst2/gitv1_e30dd01(default) srst2/gitv2_41b3f46 stacks/1.35 stacks/1.37 stacks/1.40(default) stacks/1.41 stampy/1.0.23(default) structure/2.3.4(default) tRNAscan-SE/1.3.1(default) tabix/gitv0_1ae158a tabtk/gitv0_741f3f0 tophat/2.0.10(default) treemix/gitv0_23c15a2(default) trimmomatic/0.32 trinity/2.2.0(default) trinity/20140717 use.own vcflib/gitv0_e3ab177 vcftools/0.1.11 vcftools/0.1.12a vcftools/gitv0_4fce995(default) vdb/gitv0_f1497a1 velvet/1.2.10(default) velvet/gitv0_9adf09f vsearch/gitv0_174f810(default)