CallSNPs.py
Contents
- 1 Introduction
- 2 Details of pipeline stages
- 2.1 Layout
- 2.2 Description
- 2.2.1 Calculate likelihoods of each possible variant, and apply prior and call variants
- 2.2.2 Filter variants through a maximum read depth
- 2.2.3 Housekeeping: compress vcf files
- 2.2.4 Extract features with minimum coverage
- 2.2.5 Intersect samples' features to find common feature-set
- 2.2.6 Recode vcf files to reflect common feature set
- 2.2.7 Filter recoded VCF file by quality specification
- 2.2.8 Merge VCF files
- 2.2.9 Create fasta file from merged VCF file
Introduction
The call SNPS.py script is part of the script_tools environmental module, and is loaded with:
module load script_tools
As input, it requires an alignment in the shape of the base reference sequence and the bam files of the short reads aligned to that sequence. So clearly that step should have already been taken before using CallSNPs.py. The recommended tools for that are bwa and bowtie.
Details of pipeline stages
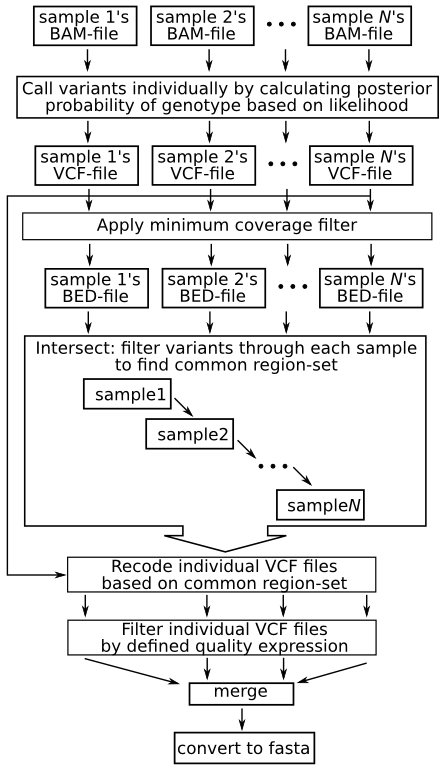
Layout
The following images summarizes the pipeline. A detailed explanation then follows:
Description
Calculate likelihoods of each possible variant, and apply prior and call variants
samtool mpileup -E -d NUM -DSugB -Q 1 -f ref_file input_bam_file | bcftools view -cgbu - > output_bcf_file
mpileup collects summary information from the input BAMs and then computes and stores the likelihood of data given each possible genotype. bcftools applies to prior and then calls the variants. Can be quite raw so often a filter is needed after this.
Filter variants through a maximum read depth
bcftools view bcf_file | vcfutils.pl varFilter -DNUM
bcftools view outputs the variants in VCF format and vcfutils.pl perl script , which is part of samtools platform, filters for variants on their maximum value via the -D option.
Housekeeping: compress vcf files
Not strictly part of the sequential run of the pipeline, but at this point the vcf files are compressed with samtools' bgzip and then indexed with tabix (also from samtools, a generic tab-file delimited file indexer).
bgzip -c vcf_file > vcf_file.gz tabix -p vcf vcf_file.gz
The latter command creates an extra vcf_file.gz.tbi file in the directory.
Extract features with minimum coverage
bedtools genomecov -bga -ibam input_bam_file | awk -v t=NUM "{if($4>=t)print}" | bedtools merge -i - > output_bed_file
bedtools genomecov outputs the number of aligned short-read bases in the genome in a format (enabled by the -bga option) in the fourth column. This is picked up by the awk program which ignores all those less than NUM. If NUM is set to zero, the entire variant callset will be returned in raw form because nothing will be filtered and the -bga (with -a presumably standing for all) will include areas with zero coverage (this sounds uninteresting but it could be useful to have, so it's often kept). The default value for NUM set in the overall python script is 15. The final merge, as well as merging the filtered output, also makes sure that split and overlapping features are rolled into one. Note that at this point our variant format VCF turns into a BED file, and we move from talking about variants to talking about regions or features, which is what bed files record.
Intersect samples' features to find common feature-set
With every short read alignment (i.e. BAM file) now having its set of features in a corresponding BED file, we look to find the features that all samples share by using bedtools' intersect subcommand. So for each sample's BED file, the following is executed>
bedtools intersect -a cumulative_intersect_bed_file -b sample_bed_file > single_intersect_bed_file mv single_intersect_file cumulative_intersect_file
At the end, the cumulative intersect file is renamed so we can know its work is finished.
mv cumulative_intersect_bed_file final_intersect_bed_file
Recode vcf files to reflect common feature set
Next we want to merge the vcf files together but taking into account our newly create intersect feature-set. This we can do with the vcftools program suite, which allows us incorporate our final intersect bed file from the last stage to produce a 'recoded' or filtered vcf for each of the samples which only includes common variants.
vcftools --gzvcf sample_vc_file --bed final_intersect_bed_file --out merged_vcf_file --recode --keep-INFO-all
This can all be done on the compressed vcf files. Once each individual sample's vcf file has been recoded in this way, we can then proceed to merge the vcf files into one.
Filter recoded VCF file by quality specification
There are certain quality consideration we want the VCF files to take into account, which samtools' bcftools filter command can achieve for us. The quality specification or expression involves merging different aspects of its value via the logical OR operation, in the shapes of i double-bar: ||:
VCF_QUALITY_EXPRESSION = '%TYPE="indel" || %TYPE="mnp" || %TYPE="other" || %QUAL<50 || INFO/DP<4 || INFO/DP4[3]<2 || INFO/DP4[4]<2 || INFO/MQ<30 || INFO/AF1<0.95 || INFO/PV4[0]<0.001 || INFO/PV4[2]<0.001 || INFO/PV4[3]<0.001'
This can be inserted into the bcftools' command-line as follows:
bcftools filter -e VCF_QUALITY_EXPRESSION sample_vcf_file | bgzip -c > compressed_sample_vcf_file
Note how the vcf output is also compressed and can also be indexed:
tabix compressed_sample_vcf_file
Merge VCF files
The vcftools program suite has a simple command for merging compressed vcfs, although its output is uncompressed, so we need to compress it also via a unix pipe (the bar |)
vcf-merge all_compressed_vcf_files | bgzip -c > merged_vcf_file_compressed
Create fasta file from merged VCF file
First however we need to render the VCF file into tabbed format, for which vcftools has the vcf-to-tab subcommand:
zcat merged_vcf_file_compressed | vcf-to-tab > merged_tabbed_vcf_file
To convert to fasta format, there is a perl script:
vcf_tab_to_fasta_alignment.pl --exclude_het -i merged_tabbed_vcf_file > merged_common_variants_fasta
The ID lines of the resulting fasta file needs to be cleaned a little to ensure later downstream compatibility.